
KDELR2 knockdown synergizes with temozolomide to induce glioma cell apoptosis through the CHOP and JNK/p38 pathways
Introduction
Glioma as the most common primary malignant brain tumors, accounts for about 30% of brain and central nervous system tumors and 80% of malignant brain tumors (1,2). Glioblastoma is the most common glioma (3). The root cause of the poor therapeutic effect of glioma is that completely removing the neoplasms is almost impossible because of the invasive growth pattern while protecting brain function. Another important reason for the poor therapeutic effect is that many chemotherapy agents penetrate the blood-brain barrier difficultly. Current treatment protocols include surgical resection, radiation therapies, and chemotherapy. At present, targeted drugs is a hot spot research in glioma therapy. Bevacizumab is the only effective targeted drug, which was proved that it prolonged progression-free in the treatment of glioma. Treatment with the chemotherapy agent temozolomide (TMZ) prolonged the median survival time by only 14.6 months in patients diagnosed with glioblastoma (4). In addition, glioma cells often acquire resistance to these agents, typically through unmethylation of MGMT promoter (4). Therefore, the identification of molecular pathways as targets of anti-glioma agents to develop novel therapies is urgently needed.
When a large number of misfolded proteins accumulate in the endoplasmic reticulum (ER), ER stress (ERS) is induced and causing changes in normal physiological conditions in the cell (5-7). To cope with ERS, chaperone proteins are released and unfolded protein response (UPR) is activated, leading to phosphorylation of protein kinase R-like ER kinase (PERK) and inositol-requiring enzyme 1 (IRE1), and activation of activating transcription factor 6 (ATF-6) (8), which further regulate associated proteins and pathways (9). A variety of solid tumors have been shown to exhibit ERS, including glioma (10); ERS is implicated in pathogenesis and apoptosis, and sustained ERS can trigger cellular death (11,12). Activation of the UPR occurs in gliomas and other human tumors (13). If ER stress exceeds the folding capacity in the ER, or defects occur in the UPR, cells are destroyed by apoptosis (8). The presence of ERS and its downstream signaling pathways also play an important role in TMZ sensitivity (14-16). Until now, the most accepted components mediating cell death under ERS downstream to PERK include the upregulation of C/EBP homologous protein (CHOP) and activating transcription factor 4 (ATF-4) (17,18). It has been reported that ERS activates the CHOP-dependent pathway that induces glioma cell apoptosis and thus the sensitivity to TMZ (15). These findings indicate that the ERS-dependent CHOP pathway and TMZ sensitivity in gliomas are closely related.
The C-terminal tetrapeptide Lys-Asp-Glu-Leu receptors (KDELRs), which regulate the retention of soluble ER molecules and vesicle trafficking, are represented by 3 members (KDELR1, KDELR2, and KDELR3) that share structural homology but have different ligands (19). When KDELR is bound with its ligands, it induces signaling cascades regulating cellular secretory traffic (20), growth, differentiation, and immune responses (21). In addition, prior research has revealed that the KDELRs participate in the ERS response (21,22), promote the recovery of misfolded proteins, enhance the adaptability of cells to survive in harmful microenvironments (23), and activate the UPR (24-26). Recent research has shown that KDELR promotes cell survival via stress-dependent activation of p38 mitogen-activated protein kinases (MAPK) (21).
The functions of KDELRs are associated with cancer biology (19,27-30), especially in glioma: KDELR1 can significantly reduce the expression of the ER transcription factor CREB3 (27) and block hypoxia-induced autophagy in glioblastoma (28); KDELR2 has been associated with the median survival rate of non-small cell lung cancer (29), and it promotes glioblastoma tumorigenesis, which is regulated by hypoxia-inducible factor 1 (HIF-1) (30). Knockdown of KDELR2 has been shown to reduce cell viability, promote cell cycle arrest, and induce apoptotic cell death by targeting cell cycle proteins (31) In addition, KDELR3 promotes high melanoma metastasis (19). The mechanism of KDELR2 action in glioma cells remains unclear, especially during response to chemotherapy and sensitivity to TMZ. In this study, we performed molecular analyses of glioma cells and investigated the effects of KDELR2 knockdown on apoptosis and the chemosensitivity to TMZ with special focus on the ERS-dependent CHOP pathway and the c-Jun amino-terminal kinase (JNK)/p38 pathway. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-869).
Methods
Antibodies and reagents
The antibodies against PERK (CST#5683), phospho-PERK (#3179), eIF2α (CST#5324), phospho-eIF2α (CST#3398), ATF-6 (#65880), XBP-1S (CST#27901), cleaved PARP (CST#5625), PARP (CST#9532), caspase 3 (CST#9661), CHOP (CST#2895), p38 (CST#9212), phospho-p38 (CST#9211), JNK (CST#9252), and phospho-JNK (CST#4668) were purchased from Cell Signaling Technology (Danvers, MA, USA). The antibodies against KDELR2 for western blotting (#ab199689), ATF-4 (#ab186284), cleaved caspase 3 (#ab90437), Bax (#ab32503), Bcl-2 (#ab194583), GRP78 (#ab21685), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (#ab181602) were purchased from Abcam (Cambridge, UK). The antibody against KDELR2 (bs-16941R) for immunohistochemistry (IHC) were purchased from Bioss Biotechnology Co., Ltd (Beijing, China). Goat anti-mouse IgG (Thermo Pierce #31160) and goat anti-rabbit IgG secondary antibody (Thermo Pierce #31210) were purchased from Thermo Fisher Scientific (Grand Island, NY, USA).
Dulbecco’s modified Eagle medium (DMEM) was obtained from Thermo Fisher Scientific. Phosphate-buffered saline (PBS) was obtained from Ausbian (Ausbian, Australia). Dimethyl sulfoxide (DMSO) and TMZ were obtained from Sigma (St. Louis, MO, USA). Cell extraction buffer was obtained from BioSource International (Camarillo, CA, USA). Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Molecular Technology (Dojindo Laboratories, Kumamoto, Japan). An annexin V-FITC/PI apoptosis detection kit was obtained from Pharmingen (San Diego, CA, USA). Polyvinylidene difluoride (PVDF) membranes were obtained from Millipore (Billerica, MA, USA). Enhanced chemiluminescence western blot reagents were obtained from Pierce Biotechnology (Rockford, IL, USA).
Histology and immunohistochemistry
To analyze the expression of KDELR2, paraffin-embedded tissue sections of tumor tissues from patients diagnosed with glioma were obtained from the Department of Neurosurgery at the Affiliated Jiujiang Hospital of Nanchang University (Jiujiang, Jiangxi, China). Glioma samples from 42 patients were obtained from 1 January 2016 to 1 September 2020; the relevant participant details are presented in Table 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All procedures were approved by the Ethics Committee of Affiliated Jiujiang Hospital of Nanchang University (JJSDYRMYY-YXLL-2020-147), and informed consent was provided by all participants.
Table 1
| Clinical character | Total cases (n=42) | Over-expression | Low-expression | P value |
|---|---|---|---|---|
| Gender (male:female) | 23:19 | 16:13 | 7:6 | 1.0 |
| Age (mean ± SD) | 47.14±8.74 | 47.38±9.21 | 46.62±7.92 | 0.786 |
| Histological diagnosis | 0.003 | |||
| Astrocytoma | 14 | 5 | 9 | |
| Glioblastoma | 28 | 24 | 4 | |
| Histological grade | 0.016 | |||
| LGG | 9 | 3 | 6 | |
| HGG | 33 | 26 | 7 |
LGG, low-grade glioma; HGG, high-grade glioma.
For IHC analysis, glioma tissue sections were generated using polyformalin-fixed and paraffin-embedded tissues as previously described (32). The tissue sections were incubated with the primary monoclonal antibodies against KDELR2. The expression levels of KDELR2 in each slide were graded according to a previously described scoring system (33). The percentage of tumor cells stained according to IHC assay were scored as 0 (no cell staining), 1 (≤30%), 2 (31–60%), or 3 (61–100%).
Cell culture
The human glioma cancer cell lines U251, U87, U373, and U118 were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS; Ausbian, Australia) and 2% penicillin/streptomycin with 5% CO2 at 37 °C.
Small interfering RNA construction and transfection
The three specific small interfering RNAs (siRNAs) for KDELR2 (siKDELR2) and negative-control scrambled siRNA were designed by GenePharma (Shanghai, China) and transfected into glioma cells using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The knockdown levels of KDELR2 were verified using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analyses. The PCR primer sequences are shown in Table 2.
Table 2
| siRNA | Sense (5'-3') | Antisense (5'-3') |
|---|---|---|
| KDELR2-1 | GCUUCUGUUUGCACUGGUCUUTT | AAGACCAGUGCAAACAGAAGCTT |
| KDELR2-2 | CAACUCGUUACCUGGAUCUUUTT | AAAGAUCCAGGUAACGAGUUGTT |
| KDELR2-3 | CCUACGAUGGAAAUCAUGAUATT | UAUCAUGAUUUCCAUCGUAGGTT |
| Scramble | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
siRNAs, small interfering RNA.
Construction of the KDELR2-overexpression vector and transfection
Full-length circular DNA (cDNA) fragments were amplified using the following primers: KDELR2F: 5'-TAGAGCTAGCGAATTCATGAACATTTTCCGGCTGACTGG-3'; KDELR2R: 5'-CTTTGTAGTCGGATCCTGCTGGCAAACTGAGCTTC-3'. After cloning into the pLenO-GTP-3XFLAG vector (Ruan Tuo, China) (34), restriction enzyme fragments were ligated into a fusion enzyme vector to construct the KDELR2 overexpression plasmids. An empty vector served as a transfection control. After transfection using Lipofectamine™ 3000 (Invitrogen, USA) according to the manufacturer’s guidelines, cells were incubated for 48 h, and then harvested. The transfection efficiency was evaluated by western blot and RT-qPCR analyses.
RT-qPCR analysis
Total RNA was prepared from cultured cells using the TRIzol® Plus RNA Purification Kit (Invitrogen, USA) following the manufacturer’s instructions. Reverse transcription assays to synthesize cDNA were performed using SuperScript™ III First-Strand Synthesis SuperMix for RT-qPCR (Invitrogen, USA). The RT-qPCR was performed using the SYBR Prellix Ex Taq™ Real-Time PCR Kit (Takara, Minato-ku, Tokyo, Japan) according to the manufacturer’s protocol. Cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 1 min. All RT-qPCR reactions were performed in triplicate. The primer sequences used for RT-qPCR are listed in Table 3. Relative quantification of the levels of each mRNA was calculated using the △△CT method and normalized to GAPDH.
Table 3
| Gene | Forward | Reverse |
|---|---|---|
| KDELR1 | AGCCACTACTTGTTTGCGCTA | CCTGCCACAATGGCGATGA |
| KDELR2 | GGAGTTTCTGGTGGTCCCTG | AATTCTGTCCGAGCACCCTG |
| KDELR3 | TCCCAGTCATTGGCCTTTCC | CCAGTTAGCCAGGTAGAGTGC |
| Atf-6 | CGAAGGGATCACCTGCTGTT | CCTGGTGTCCATCACCTGAC |
| eIF2α | TGGTGAATGTCAGATCCATTGC | TAGAACGGATACGCCTTCTGG |
| ATF4 | GTCCTGTCCTCCACTCCAGA | GGGTGTCTTCCTCCTTTATGC |
| GRP78 | CTGCTATCCAACGCACTGGT | GGTGGCCCAGAGACAGATTC |
| PERK | ACGATGAGACAGAGTTGCGAC | ATCCAAGGCAGCAATTCTCCC |
| CHOP | TCTTCCTCCTCTTCCTCCTG | CACTCTTGACCCTGCTTCTC |
| GAPDH | CCATGACAACTTTGGTATCGTGGAA | GGCCATCACGCCACAGTTTC |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
CCK-8 assay
The CCK-8 assay was performed according to the manufacturer’s instructions. Briefly, each group of U373 cells (2×103 cells per well) were seeded in 96-well plates filled with 100 µL of medium per well and cultured for the indicated times (1, 2, 3, 4, or 5 days). Then, 10 µL of CCK-8 solution was added into each well at each time point. The absorbance [optical density (OD)] in each well was estimated using a microplate reader (Bio-Rad, Hercules, CA, USA) at a wavelength of 450 nm after 4 h of incubation. The OD 450 and OD 450/fold were calculated to assess cell viability.
Flow cytometry assay
For the flow cytometry analysis of apoptosis, U373 cells (siRNA control, siKDELR2, overexpressed control, and overexpressed KDELR2) were harvested and washed with PBS twice. Then, the cells were incubated with FITC-conjugated annexin V and propidium iodide (PI), following the manufacturer’s instructions (BD, San Diego, CA, USA). Briefly, the cells were then collected and fixed overnight in 70% ethanol at 4 °C, and subsequently stained using annexin V-FITC (10 µg/mL) and PI (5 µg/mL) for 30 min at 37 °C. Samples were analyzed on an LSR II flow cytometer using CELL Quest software for acquisition and FCS Express software (De Novo Software, CA, USA) for analysis.
Hoechst 33258 nuclear staining
To assess the apoptotic effect of downregulating KDELR2 in U373 cells, apoptotic cells were analyzed using the Hoechst 33258 nuclear staining assay. Each group of U373 cells was air-dried and fixed with 4% paraformaldehyde for 30 min. Cells were then washed 3 times with PBS for 5 min. Each group of cells was then incubated with Hoechst 33258 stain at room temperature for 10 min, soaked in PBS for 5 min, and washed 3 times. The slides were sealed with an anti-fluorescence quenching liquid. The apoptotic cells exhibited intense fluorescence and chromatin condensation when observed under a fluorescence microscope.
Colony formation assay
Each group of U373 cells (500 cells per well) were seeded into 6-well plates and incubated in DMEM with 10% FBS and 2% penicillin/streptomycin for 2 weeks. The colonies were washed with PBS, fixed with methanol for 15 min, and subsequently stained with 1,000 µL 0.15% crystal violet (Sigma, San Francisco, CA, USA). The colonies were scanned and counted using a high-resolution digital camera.
Western blot assay
The glioma tissues and U373 cells of each group were harvested, and lysates of the tissues and cells were generated using radioimmunoprecipitation assay (RIPA) lysis buffer for approximately 30 min at 4 °C. Protein extracts (30 µg) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then immediately transferred onto PVDF membranes listed above, and subsequently incubated with the abovementioned corresponding primary and secondary antibodies. Membranes containing the transferred protein were blocked in 5% defatted skim milk in tris-buffered saline with Tween-20 (TBST) and incubated with a primary antibody overnight at 4 °C, followed by an incubation with secondary antibodies for 2 h at 4 °C. The signals from the antibody complexes were detected using an enhanced chemiluminescence kit (Applygen, Beijing, China).
TMZ sensitivity assay
To determine whether KDELR2 knockdown enhances TMZ sensitivity to glioma cells, the IC50 of TMZ was firstly evaluated. Briefly, U373 cells were seeded in 96-well plates and incubated in DMEM with TMZ (10, 50, 100, 200, 400, 600, 800, 1,000, and 1,200 nM, DMSO served as control) and continuously cultured for 72 h at 37 °C. Next, CCK-8 solution (10 µL) was added to the plates and the absorbance in each well was measured as listed above. The formula for the in vitro cell inhibition rate = (1-OD 450 of treatment group/OD 450 of control group) ×100%. The IC50 was calculated according to the OD 450 from the CCK-8 assays. Subsequently, the methods for assessing proliferation by CCK-8 assays and apoptosis by flow cytometry and Hoechst 33258 nuclear staining assay were performed as described above to detect if KDELR2 knockdown enhances TMZ sensitivity.
Statistical analysis
Data were presented as the mean ± standard deviation (SD) from at least three independent experiments. Statistical analyses were performed using the software SPSS 23.0 (IBM SPSS Inc., Chicago, IL, USA) and GraphPad Prism (GraphPad Inc., San Diego, CA, USA). Pearson’s χ2 test was used to analyze the association between gene expression and clinic pathological characteristics. Statistical comparisons were analyzed using one-way analysis of variance (ANOVA) followed by Dunnett’s t test. Results were considered statistically significant at P<0.05.
Results
Elevated KDELR2 expression in glioma tissues is correlated with high-grade glioma
Our research revealed that KDELR2 is overexpressed in glioblastoma tissues and may be associated with poor prognosis; however, its expression in different grades of glioma is not clear. To analyze the relative expression of KDELR2 in glioma tissues, we performed IHC staining and western blot analysis in low-grade glioma (LGG) and high-grade glioma (HGG) tissues in a cohort of 42 glioma patients (LGG: 9 cases, HGG: 33 cases). There were 29 cases with upregulated KDELR2, comprising 69.0% of the total cases. The results revealed that KDELR2 protein expression was significantly upregulated in HGG tissues (Figure 1A). Among 33 cases of HGG, there were 26 cases (78.8%) with upregulated KDELR2. As shown in Table 1, elevated KDELR2 expression was significantly correlated with HGG, as assessed by IHC, and glioblastoma. Western blot analysis confirmed the IHC results (Figure 1B). Western blot analysis was performed to analyze the relative expression of KDELR2 in paired glioma and paraglioma tissues. The results showed that the KDELR2 protein levels in glioma tissues were significantly increased compared to those in matched paraglioma tissues (Figure 1C).
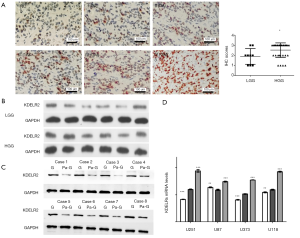
KDELR2 expression in glioma cell lines
To determine an appropriate glioma cell line, we measured the messenger RNA (mRNA) levels of the KDELRs (KDELR1, KDELR2, and KDELR3) using RT-qPCR in U251, U87, U373, and U118 cell lines (Figure 1D). According to the result, U373 cells were selected for subsequent analyses to avoid interference with higher or lower KDELR1 and KDELR3.
KDELR2 knockdown inhibits cell proliferation and induces cell apoptosis in glioma
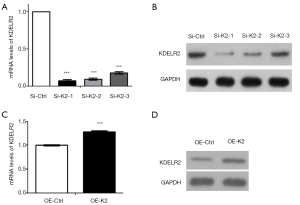
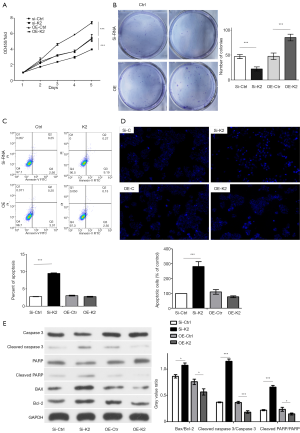
As previously reported, KDELR knockdown attenuated the anti-apoptotic effects of Santacruzamate A in the pathophysiology of Alzheimer’s disease (35), and KDELR2 knockdown reduced cell viability and inhibited colony formation in glioblastoma cells (30). To measure the effect of KDELR2 knockdown in glioma cells, we established KDELR2 knockdown U373 cells with siRNAs (siKDELR2#1, siKDELR2#2, and siKDELR2#3) evaluated them by RT-qPCR (Figure 2A) and western blot analysis (Figure 2B). KDELR2 overexpression cells was established with pLenO-K2 vector evaluated them by RT-qPCR (Figure 2C) and western blot analysis (Figure 2D). Subsequently, siKDELR2#1 was selected for subsequent studies. On the one hand, the results of CCK-8 and colony formation assays showed that KDELR2 downregulation suppressed the proliferation in U373 cells according to the decreased cell viability (Figure 3A) and colony number (Figure 3B). On the other hand, the increased apoptosis rate in the flow cytometry assay as well as the in Hoechst 33258 stain assay illustrated that KDELR2 deficiency caused a significant increase in apoptosis of U373 cells (Figure 3C,D). Conversely, overexpression of KDELR2 produced opposite effects in U373 cells (Figure 3A,B,C,D). Furthermore, western blot assay was used to measure the expression of apoptosis-related proteins. When silencing KDELR2, the levels of pro-apoptotic proteins [cleaved poly ADP ribose polymerase (PARP), cleaved caspase 3, and Bcl-2-associated X protein (Bax)] were elevated (Figure 3E).
KDELR knockdown induces apoptosis via the ERS-dependent CHOP pathway
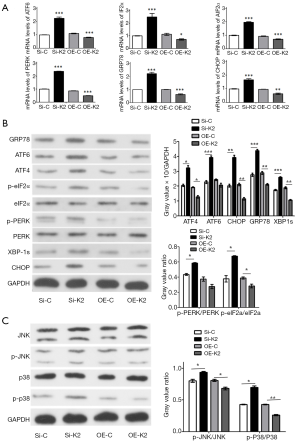
To determine whether KDELR knockdown-induced glioma cell apoptosis was associated with the ERS-dependent CHOP pathway, we focused on the KDELR2 knockdown-induced apoptotic pathway, which is associated with ERS signaling. In this study, the results of RT-qPCR demonstrated that silencing KDELR2 upregulated the levels of the ERS and UPR-related genes, including ATF6, eIF2α, ATF4, GRP78, PERK, and CHOP (Figure 4A) comparing with SiRNA control group (Si-C). However, the levels of these genes were downregulated in KDELR2 overexpressing U373 cells (Figure 4A) comparing with overexpression control group (OE-C). Collectively, these results indicated that KDELR2 knockdown induces ERS concomitant with CHOP activation. Next, the results of western blotting showed that when silencing KDELR2 in U373 cells, the expression of ATF6, ATF4, XBP1s, and CHOP were upregulated comparing with control groups (Figure 4B). We also investigated the presence of GRP78 protein, a crucial chaperone protein for KDELR2, which monitors ER protein activities. In this study, GRP78 protein was upregulated in KDELR2 knockdown cells comparing with Si-C, but downregulated in the KDELR2 overexpressed group comparing with OE-C (Figure 4B). To determine whether the UPR was activated in response to silencing KDELR2, the levels of phosphorylated PERK, which leads to the activation of eIF2α (p-eIF2α), were also detected. After silencing KDELR2, although the total expression of PERK and eIF2α was not significantly changed, PERK and eIF2α were activated (phosphorylated forms) comparing with Si-C, as shown in Figure 4B. These results suggested that the ERS-dependent CHOP pathway is one of the mechanisms by which KDELR2 knockdown induces apoptosis in U373 cells.
KDELR knockdown enhances apoptosis via the JNK/p38 pathway
The MAPK signaling pathway plays an important role in apoptosis (36). It is not only by its retrieval ability of ER molecular chaperone proteins that KDELR participates in the ERS response, but also by modulating MAPK signaling and JNKs (21). To determine whether KDELR2-downregulated apoptosis was associated with MAPK signaling, we examined whether MAPK signaling was activated during KDELR2 knockdown-induced apoptosis. Intriguingly, after silencing KDELR2, although the total expression of JNK and p38 was not affected, it upregulated JNK and p38 activation (phosphorylated forms), which were decreased in the KDELR2 overexpressing U373 cells (Figure 4C). These results revealed that the JNK/p38 signaling pathway is one of the mechanisms by which KDELR2 knockdown induces apoptosis in U373 cells.
KDELR2 knockdown sensitizes U373 cells to TMZ
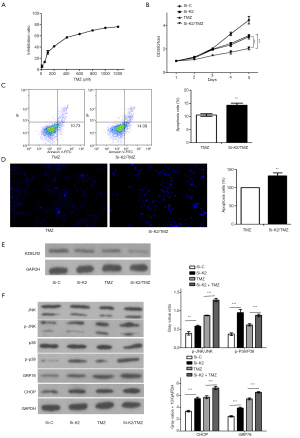
Due to its strong DNA toxicity and high penetrability to cross the blood-brain barrier, TMZ has been used in first-line chemotherapy for glioma. Both ERS and its downstream components have important roles in regulating TMZ sensitivity (8). Although some protein interactions between TMZ and the UPR have been found, the interaction between KDELR2 and TMZ, and the underlying regulatory mechanisms are still not understood. To test whether the efficacy of TMZ could be improved if used in combination with KDELR2 siRNA, we tested KDELR2 siRNA and a sub-IC50 dose of TMZ (200 µM) each alone and in combination.
As shown in Figure 5A, TMZ reduced cell viability after 5 days of treatment in a dose-dependent manner, with 5 days IC50 of TMZ of 301.3 µM [95% confidence interval (CI): 281.8 to 322.1]. The knockdown of KDELR2 showed synergy with TMZ in inhibiting cell proliferation, as measured using CCK-8 assay (Figure 5B), and showed an enhanced apoptotic ratio compared with TMZ alone by flow cytometry (Figure 5C) and Hoechst 33258 staining (Figure 5D). There was marked inhibition of KDELR2 in the KDELR2 knockdown/TMZ treatment in U373 cells compared to the individual treatments alone (Figure 5E), suggesting a potential role of KDELR2 in mediating the TMZ response. The JNK/p38 and CHOP signaling proteins and some associated molecules, such as JNK, p-JNK, p38, p-p38, GRP78, and CHOP, were increased in the KDELR2 knockdown/TMZ treatment compared to KDELR2 knockdown and TMZ alone (Figure 5F). These results suggested that reducing KDELR2 expression in U373 cells promoted its sensitivity to TMZ via the JNK/p38 and ERS-dependent CHOP pathways.
Discussion
The KDEL receptors play significant roles in ER quality control by retro-transporting ER chaperone proteins (37,38), and allow for protein cargoes to move toward the plasma membrane via anterograde transport (39). Using anti-KDEL agents is a promising tumor-targeting approach for treating ovarian cancer (40). Knockdown of KDELR1 increases metastasis (19), but knockdown of KDELR3 diminishes metastasis. Although studies have suggested that KDELR2 participates in glioblastoma progression and could be a novel therapeutic target and prognostic factor (30,31), no detailed studies on its effect on glioma cell proliferation or apoptosis have been conducted. Our findings showed that KDELR2 exerted its effects at multiple levels, suggesting additional unidentified roles. In this study, knockdown of KDELR2 inhibited proliferation, promoted apoptosis, and increased the sensitivity of chemotherapy to TMZ in U373 cells by activating the ERS-dependent CHOP pathway and via the JNK/p38 pathway.
Knockdown of KDELR2 induced significant apoptosis in glioma cells, which suggested that silencing KDELR2-reduced glioma growth relies predominantly on the induction of apoptosis. Whether ERS plays an important role in silencing KDELR2-induced glioma cell apoptosis remains uncertain. The KDELR-mediated retrieval of ER molecular chaperone proteins is limited by the expression of a mutant KDEL receptor, which dramatically aggravates ERS (35,41). It has been shown that KDELR2 interacts with GRP78 (26), and the expression of GRP78, which regulates the UPR in the ER, is upregulated when ERS occurs (25). Aggregation of misfolded and unfolded proteins in the ER lumen induces GRP78 release from the lumenal domains of ER receptors, which facilitates proper protein folding and assembly (42). It was previously shown that upregulation of GRP78 reinforces ER homeostasis and the UPR and ERS-induced apoptosis in malignant glioma cells (24,25,43). Our results suggested that knockdown of KDELR2 in U373 cells significantly elevated levels of GRP78, XBP1s, and ATF6, which are all ERS-related proteins. Our results further suggested that knockdown of KDELR2 aggravated ERS.
It has been shown that ERS induces KDELRs binding to ERS sensors PERK, IRE1, and ATF6 and activates UPR pathways (24-26), including the PERK-eIF2α-ATF4-CHOP major pathway (44). The activation of PERK signaling has been shown to impair cell proliferation and promote apoptosis (45). Our results suggested that knockdown of KDELR2 significantly elevated the levels of phosphorylated PERK, which is consistent with the findings of a previous study (35). Activation of PERK results in subsequent phosphorylation of eIF2, causing signal switching to enhance apoptotic cell death by causing increased translation of ATF-4 (46), which promotes the expression of transcription factors including CHOP, a central factor in cell apoptosis (24,47-49). As shown in Figure 4, knockdown of KDELR2 increased the mRNA levels of eIF2α, ATF4, PERK, and CHOP, and increased the levels of p-PERK, p-eIF2α, and CHOP. Furthermore, silencing KDELR2 upregulated ATF6 expression, which might have led to a further increase in CHOP. It has been shown that CHOP directly regulates death effectors such as Bcl-2 (50) and activates caspase-3 (51). We found that the ratio of Bax/Bcl-2 and expression of Bax was increased in KDELR2 knockdown cells. In addition, cleaved caspase-3 was upregulated, suggesting that caspase-3 was also activated by KDELR2 knockdown. These effects could be rescued by KDELR2 overexpression. These results suggested that KDELR2 knockdown might also induce apoptosis in chondrocytes via caspase activation, and that knockdown of KDELR2 induces apoptosis via the ERS-dependent CHOP pathway.
Recent findings have suggested that KDELR activates p38 MAPK (21). Under ERS, cells activate JNK and p38 MAPK, which contributes to apoptosis (52-54). Endothelial cell apoptosis is mediated by ERS via activation of the JNK/p38-dependent pathway (54). As shown in Figure 5, KDELR2 knockdown did not increase the protein levels of IRE1α, JNK, and p38; however, it increased p-IRE1α, p-JNK, and p-p38 levels. These results suggested that knockdown of KDELR2 may induce apoptosis via the JNK/p38 pathway. In addition, a recent study showed that patterns of apoptotic activation from stress signaling were different from those of ERS, and that apoptotic activation at moderate levels of stress is dependent upon PERK and p38 signaling (55). Therefore, further research is required to evaluate the degree of ERS caused by KDELR2 knockdown.
There is an urgent need to improve the current chemotherapy available to glioma patients. Identifying the novel mechanisms of TMZ resistance, overcoming TMZ chemoresistance, and developing new agents need to be improved. There is potential for ERS as a synergistic cytosolic target of TMZ-induced apoptosis (56). It has been shown that TMZ itself can lead to activation of the UPR (57) and induce GRP78 and CHOP in glioma cells, which decisively contribute to the phenotypic outcome and determine the balance between subsequent cell survival and cell death (56,58). Activation of CHOP might enhance the sensitivity of glioma cells to TMZ (15). The ERS-inducing agent JLK1486 combined with TMZ has been shown to decrease cell growth and increase cell death in glioblastoma cell lines, mediated by increasing levels of ATF4 and CHOP (59). These results suggest a close relationship between ERS and the TMZ sensitivity of gliomas.
We observed for the first time that KDELR2 was involved in TMZ-induced apoptosis, and that knockdown of KDELR2 significantly increased the sensitivity of U373 cells to TMZ. In addition, we found that TMZ decreased KDELR2 expression in U373 cells with silencing of KDELR2. Furthermore, the KDELR2 knockdown/TMZ treatment increased the levels of p-JNK and p-p38, and particularly increased the levels of CHOP in the ERS-dependent CHOP pathway. Taken together, our data suggest that KDELR2 knockdown and TMZ combination therapy inhibits proliferation and induces apoptosis via the ERS-dependent CHOP and JNK/p38 pathways in glioma cells, but further studies are needed to elucidate its precise mechanism of action. Above all, our investigation has highlighted the potential for combined use of TMZ and silencing of KDELR2 in the treatment of glioma.
To summarize, our research showed that knockdown of KDELR2 is a potential strategy to treat glioma, and that it exerted a synergistic effect on glioma cells when combined with TMZ via the ERS-dependent CHOP and JNK/p38 pathways. Further studies are needed to elucidate the mechanism of the related apoptotic pathway and the synergistic effect between knockdown of KDELR2 and TMZ action.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-21-869
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-869
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-869). All authors report funding from Science and Technology Research Project of Education Department of Jiangxi Province (No. 170101). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All procedures were approved by the Ethics Committee of Affiliated Jiujiang Hospital of Nanchang University (JJSDYRMYY-YXLL-2020-147), and informed consent was provided by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol 2014;16:896-913. [Crossref] [PubMed]
- Johnson GG, White MC, Grimaldi M. Stressed to death: targeting endoplasmic reticulum stress response induced apoptosis in gliomas. Curr Pharm Des 2011;17:284-92. [Crossref] [PubMed]
- Global Burden of Disease Cancer Collaboration. The Global Burden of Cancer 2013. JAMA Oncol 2015;1:505-27. [Crossref] [PubMed]
- Tremont-Lukats IW, Teh BS. Lomustine and temozolomide for newly diagnosed glioblastoma with methylated MGMT promoter: Lessons from the CeTeG/NOA-09 trial. Transl Cancer Res 2019;8:S589-91. [Crossref]
- Xu K, Han B, Bai Y, et al. MiR-451a suppressing BAP31 can inhibit proliferation and increase apoptosis through inducing ER stress in colorectal cancer. Cell Death Dis 2019;10:152. [Crossref] [PubMed]
- Wu HL, Duan ZT, Jiang ZD, et al. Increased endoplasmic reticulum stress response is involved in clopidogrel-induced apoptosis of gastric epithelial cells. PLoS One 2013;8:e74381. [Crossref] [PubMed]
- Hou X, Fu M, Cheng B, et al. Galanthamine improves myocardial ischemia-reperfusion-induced cardiac dysfunction, endoplasmic reticulum stress-related apoptosis, and myocardial fibrosis by suppressing AMPK/Nrf2 pathway in rats. Ann Transl Med 2019;7:634. [Crossref] [PubMed]
- He Y, Su J, Lan B, et al. Targeting off-target effects: endoplasmic reticulum stress and autophagy as effective strategies to enhance temozolomide treatment. Onco Targets Ther 2019;12:1857-65. [Crossref] [PubMed]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012;13:89-102. [Crossref] [PubMed]
- Wang J, Qi Q, Zhou W, et al. Inhibition of glioma growth by flavokawain B is mediated through endoplasmic reticulum stress induced autophagy. Autophagy 2018;14:2007-22. [Crossref] [PubMed]
- Limonta P, Moretti RM, Marzagalli M, et al. Role of Endoplasmic Reticulum Stress in the Anticancer Activity of Natural Compounds. Int J Mol Sci 2019;20:961. [Crossref] [PubMed]
- Chang CY, Li JR, Wu CC, et al. Endoplasmic Reticulum Stress Contributes to Indomethacin-Induced Glioma Apoptosis. Int J Mol Sci 2020;21:557. [Crossref] [PubMed]
- Clarke HJ, Chambers JE, Liniker E, et al. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014;25:563-73. [Crossref] [PubMed]
- Peñaranda Fajardo NM, Meijer C, Kruyt FA. The endoplasmic reticulum stress/unfolded protein response in gliomagenesis, tumor progression and as a therapeutic target in glioblastoma. Biochem Pharmacol 2016;118:1-8. [Crossref] [PubMed]
- Ma J, Yang YR, Chen W, et al. Fluoxetine synergizes with temozolomide to induce the CHOP-dependent endoplasmic reticulum stress-related apoptosis pathway in glioma cells. Oncol Rep 2016;36:676-84. [Crossref] [PubMed]
- Sun Y, Zhang X. Bufothionine Promotes Apoptosis via Triggering ER Stress and Synergizes with Temozolomide in Glioblastoma Multiforme Cells. Anat Rec (Hoboken) 2019;302:1950-7. [Crossref] [PubMed]
- Puthalakath H, O'Reilly LA, Gunn P, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 2007;129:1337-49. [Crossref] [PubMed]
- Urra H, Dufey E, Lisbona F, et al. When ER stress reaches a dead end. Biochim Biophys Acta 2013;1833:3507-17. [Crossref] [PubMed]
- Marie KL, Sassano A, Yang HH, et al. Melanoblast transcriptome analysis reveals pathways promoting melanoma metastasis. Nat Commun 2020;11:333. [Crossref] [PubMed]
- Giannotta M, Ruggiero C, Grossi M, et al. The KDEL receptor couples to Gαq/11 to activate Src kinases and regulate transport through the Golgi. EMBO J 2012;31:2869-81. [Crossref] [PubMed]
- Yamamoto K, Hamada H, Shinkai H, et al. The KDEL receptor modulates the endoplasmic reticulum stress response through mitogen-activated protein kinase signaling cascades. J Biol Chem 2003;278:34525-32. [Crossref] [PubMed]
- Wang P, Li B, Zhou L, et al. The KDEL receptor induces autophagy to promote the clearance of neurodegenerative disease-related proteins. Neuroscience 2011;190:43-55. [Crossref] [PubMed]
- Trychta KA, Bäck S, Henderson MJ, et al. KDEL Receptors Are Differentially Regulated to Maintain the ER Proteome under Calcium Deficiency. Cell Rep 2018;25:1829-1840.e6. [Crossref] [PubMed]
- Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013;32:805-18. [Crossref] [PubMed]
- Hammadi M, Oulidi A, Gackière F, et al. Modulation of ER stress and apoptosis by endoplasmic reticulum calcium leak via translocon during unfolded protein response: involvement of GRP78. FASEB J 2013;27:1600-9. [Crossref] [PubMed]
- Tiwarekar V, Fehrholz M, Schneider-Schaulies J. KDELR2 Competes with Measles Virus Envelope Proteins for Cellular Chaperones Reducing Their Chaperone-Mediated Cell Surface Transport. Viruses 2019;11:27. [Crossref] [PubMed]
- Oh-Hashi K, Soga A, Naruse Y, et al. Elucidating post-translational regulation of mouse CREB3 in Neuro2a cells. Mol Cell Biochem 2018;448:287-97. [Crossref] [PubMed]
- Xue H, Zhang J, Guo X, et al. CREBRF is a potent tumor suppressor of glioblastoma by blocking hypoxia-induced autophagy via the CREB3/ATG5 pathway. Int J Oncol 2016;49:519-28. [Crossref] [PubMed]
- Bajaj R, Kundu ST, Grzeskowiak CL, et al. IMPAD1 and KDELR2 drive invasion and metastasis by enhancing Golgi-mediated secretion. Oncogene 2020;39:5979-94. [Crossref] [PubMed]
- Liao Z, She C, Ma L, et al. KDELR2 Promotes Glioblastoma Tumorigenesis Targeted by HIF1a via mTOR Signaling Pathway. Cell Mol Neurobiol 2019;39:1207-15. [Crossref] [PubMed]
- Mao H, Nian J, Wang Z, et al. KDELR2 is an unfavorable prognostic biomarker and regulates CCND1 to promote tumor progression in glioma. Pathol Res Pract 2020;216:152996. [Crossref] [PubMed]
- Wang X, Tao C, Yuan C, et al. AQP3 small interfering RNA and PLD2 small interfering RNA inhibit the proliferation and promote the apoptosis of squamous cell carcinoma. Mol Med Rep 2017;16:1964-72. [Crossref] [PubMed]
- Huang W, Zhong Z, Luo C, et al. The miR-26a/AP-2α/Nanog signaling axis mediates stem cell self-renewal and temozolomide resistance in glioma. Theranostics 2019;9:5497-516. [Crossref] [PubMed]
- Chen Y, Chen X, Ji YR, et al. PLK1 regulates hepatic stellate cell activation and liver fibrosis through Wnt/β-catenin signalling pathway. J Cell Mol Med 2020;24:7405-16. [Crossref] [PubMed]
- Chen L, Liu YC, Tan H, et al. Santacruzamate A Ameliorates AD-Like Pathology by Enhancing ER Stress Tolerance Through Regulating the Functions of KDELR and Mia40-ALR in vivo and in vitro. Front Cell Neurosci 2019;13:61. [Crossref] [PubMed]
- Ou YW, Zhao ZT, Wu CY, et al. Mig-2 attenuates cisplatin-induced apoptosis of human glioma cells in vitro through AKT/JNK and AKT/p38 signaling pathways. Acta Pharmacol Sin 2014;35:1199-206. [Crossref] [PubMed]
- Wiersma VR, Michalak M, Abdullah TM, et al. Mechanisms of Translocation of ER Chaperones to the Cell Surface and Immunomodulatory Roles in Cancer and Autoimmunity. Front Oncol 2015;5:7. [Crossref] [PubMed]
- Zhang C, Syed TW, Liu R, et al. Role of Endoplasmic Reticulum Stress, Autophagy, and Inflammation in Cardiovascular Disease. Front Cardiovasc Med 2017;4:29. [Crossref] [PubMed]
- Cancino J, Jung JE, Luini A. Regulation of Golgi signaling and trafficking by the KDEL receptor. Histochem Cell Biol 2013;140:395-405. [Crossref] [PubMed]
- Delie F, Ribaux P, Petignat P, et al. Anti-KDEL-coated nanoparticles: a promising tumor targeting approach for ovarian cancer? Biochimie 2012;94:2391-7. [Crossref] [PubMed]
- Blum A, Khalifa S, Nordström K, et al. Transcriptomics of a KDELR1 knockout cell line reveals modulated cell adhesion properties. Sci Rep 2019;9:10611. [Crossref] [PubMed]
- Dandekar A, Mendez R, Zhang K. Cross talk between ER stress, oxidative stress, and inflammation in health and disease. Methods Mol Biol 2015;1292:205-14. [Crossref] [PubMed]
- Markouli M, Strepkos D, Papavassiliou AG, et al. Targeting of endoplasmic reticulum (ER) stress in gliomas. Pharmacol Res 2020;157:104823. [Crossref] [PubMed]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011;13:184-90. [Crossref] [PubMed]
- Lin JH, Li H, Zhang Y, et al. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One 2009;4:e4170. [Crossref] [PubMed]
- Han J, Back SH, Hur J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013;15:481-90. [Crossref] [PubMed]
- Hu J, Dang N, Menu E, et al. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood 2012;119:826-37. [Crossref] [PubMed]
- Teske BF, Wek SA, Bunpo P, et al. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol Biol Cell 2011;22:4390-405. [Crossref] [PubMed]
- Wu YJ, Su TR, Dai GF, et al. Flaccidoxide-13-Acetate-Induced Apoptosis in Human Bladder Cancer Cells is through Activation of p38/JNK, Mitochondrial Dysfunction, and Endoplasmic Reticulum Stress Regulated Pathway. Mar Drugs 2019;17:287. [Crossref] [PubMed]
- Scull CM, Tabas I. Mechanisms of ER stress-induced apoptosis in atherosclerosis. Arterioscler Thromb Vasc Biol 2011;31:2792-7. [Crossref] [PubMed]
- Uehara Y, Hirose J, Yamabe S, et al. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthritis Cartilage 2014;22:1007-17. [Crossref] [PubMed]
- Hetz C, Bernasconi P, Fisher J, et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 2006;312:572-6. [Crossref] [PubMed]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011;334:1081-6. [Crossref] [PubMed]
- Legeay S, Fautrat P, Norman JB, et al. Selective deficiency in endothelial PTP1B protects from diabetes and endoplasmic reticulum stress-associated endothelial dysfunction via preventing endothelial cell apoptosis. Biomed Pharmacother 2020;127:110200. [Crossref] [PubMed]
- Lumley EC, Osborn AR, Scott JE, et al. Moderate endoplasmic reticulum stress activates a PERK and p38-dependent apoptosis. Cell Stress Chaperones 2017;22:43-54. [Crossref] [PubMed]
- Chen TC, Wang W, Golden EB, et al. Green tea epigallocatechin gallate enhances therapeutic efficacy of temozolomide in orthotopic mouse glioblastoma models. Cancer Lett 2011;302:100-8. [Crossref] [PubMed]
- Pyrko P, Schönthal AH, Hofman FM, et al. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res 2007;67:9809-16. [Crossref] [PubMed]
- Schönthal AH. Endoplasmic reticulum stress and autophagy as targets for cancer therapy. Cancer Lett 2009;275:163-9. [Crossref] [PubMed]
- Weatherbee JL, Kraus JL, Ross AH. ER stress in temozolomide-treated glioblastomas interferes with DNA repair and induces apoptosis. Oncotarget 2016;7:43820-34. [Crossref] [PubMed]
(English Language Editor: J. Jones)


