
Systematically integrative analysis identifies diagnostic and prognostic candidates and small-molecule drugs for lung adenocarcinoma
Introduction
Lung cancer (LC), one of the most common malignancies, is the leading cause of cancer-related deaths worldwide (1).Especially in recent years, the morbidity and mortality of LC have been increasing year by year, and LC has ranked first among all malignancies for many years in some countries such as China (2019 National Cancer Report from China National Cancer Center). Non-small cell lung cancer (NSCLC) is the most predominant pathological type, constituting more than 80% of LCs (2), of which lung adenocarcinoma (LUAD) is the major histological subtype and accounts for more than 40% of LCs (3,4). Annually, LUAD results in more than 600,000 deaths all over the world (5). Despite recent advances in molecular diagnosis and multimodality therapies, the 5-year overall survival (OS) rate of LUAD patients in all stages is only approximately 15% (3,6). LUAD patients diagnosed at an early stage have a higher 5-year OS rate with 70–90% (7). However, no more than 20% of LUAD patients are diagnosed in a timely manner at an early stage (8), and 35–50% of patients diagnosed and treated at an early stage will relapse after surgical resection (9), which indicates a very poor prognosis. To improve the survival rate of LUAD patients, it is vital to uncover the underlying molecular mechanisms of LUAD and identify potential molecular diagnostic and prognostic biomarkers and/or therapeutic targets to combat LUAD.
Currently, the diagnosis and prognosis of LUAD patients are mainly evaluated on the basis of many clinical and pathological features. Due to the high heterogeneity of LUAD, many clinical variables correlating with prognosis bring difficulty in predicting clinical outcomes upon detecting LUAD at an early stage (10). Recently, several molecular factors such as gene mutation and overexpression have been used to guide the clinical care of LUAD patients (3,11). For example, EGFR mutations and ALK fusions have been used as targets of molecular targeted therapy (3,12,13). However, EGFR and ALK alterations were found in only a small fraction of LUAD patients, and the majority of LUAD patients frequently harbored activating mutations such as KRAS, BRAF and ERBB2 (14) and loss-of-function mutations and deletions such as TP53, RB1 and CDKN2A (3,15,16). So far, few targeted molecular therapies have been clinically used for such alterations, and few prognostic biomarkers have been identified to predict clinical outcomes. Therefore, more knowledge of additional genes altered in LUAD is required to further guide the diagnosis, treatment and prognosis of LUAD.
Gene expression analysis based on gene expression profiles is an important traditional method of investigating the differences in gene expression under different cell statuses, and a large number of differentially expressed genes (DEGs) associated with LUAD have been identified (17-20), such as AKT1, DDR2 and FGFR1. Some genetic factors including K-ras, FGF22 and LAPTM4B, as biomarkers, have also been investigated to predict the prognosis in LUAD patients (21-23). However, most DEGs reported in different studies vary greatly, and only a few consistent DEGs have been identified. In addition, few identified diagnostic and prognostic markers have been widely accepted for routine clinical use, and widely acceptable consistent key genes as biomarkers urgently require identification.
Increasing LUAD-related gene expression data allows us to do the important work of identifying consistent key genes involved in LUAD by systematically integrative analysis. In this study, five LUAD-related gene expression profile datasets from the NCBI Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases were systematically integrated by bioinformatics methods including weighted gene coexpression network analysis (WGCNA), differentially expressed gene analysis (DEGA), functional enrichment analysis, and protein and protein interaction (PPI) network construction. Subsequently, diagnostic and prognostic analyses of identified critical genes were performed to identify diagnostic and prognostic candidates associated with LUAD patients. Last, potential small-molecule drugs related to key genes were identified to guide the treatment of LUAD.
We present the following article in accordance with the TRIPOD reporting checklist and the MDAR checklist (available at https://dx.doi.org/10.21037/tcr-21-526).
Methods
The flow chart of systematic bioinformatics analysis in the current study is displayed in Figure 1.
Data collection
Five LUAD-related gene expression datasets were reanalyzed by systematic bioinformatics methods in this study. Among these datasets, four microarray datasets were retrieved from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geoprofiles/), including GSE10072 (19), GSE7670 (24), GSE19804 (20) and GSE102511 (25). The GSE10072, GSE7670 and GSE19804 datasets were generated using the Affymetrix microarray platform. The GSE102511 dataset was produced using the Ion Torrent Proton platform. The GSE10072 and GSE102511 datasets were from American patients with LUAD. The GSE10072 dataset included 58 eligible tumor tissues samples and 49 eligible normal lung tissues samples, and the GSE102511 dataset included 16 eligible tumor tissues samples and 15 eligible normal lung tissues samples. The GSE7670 and GSE19804 datasets were from Taiwanese patients with LUAD and included 27 and 56 eligible paired tissue samples, separately. Given the same microarray chip platform (GPL96), the GSE10072 and GSE7670 datasets were merged into one dataset by reconstructing the gene expression profiles, and the new dataset was named “Dataset A”. The GSE19804 and GSE102511 datasets were named “Dataset B” and “Dataset C”, respectively.
A LUAD-related RNA-seq dataset was retrieved from TCGA database portal (2019, https://portal.gdc.cancer.gov/). The inclusion criteria for the RNA-seq data were as follows: (I) histological diagnosis for LUAD; (II) except LUAD, not suffering from other malignancies; and (III) data with complete clinical information. Finally, totals of 535 LUAD tissues samples and 59 non-LUAD normal lung tissues samples were included. The LUAD-related RNA-seq dataset was named “Dataset D”.
These studies have been approved by the Institutional Review Board of the relevant participating institutions including the National Taiwan University and Taichung Veterans General Hospital (GSE19804), the Taipei Veterans General Hospital and Taichung Veterans General Hospital (GSE7670), 13 participating hospitals and National Cancer Institutes (GSE10072), the Aichi Cancer Center and Nagasaki University (GSE102511), and the National Cancer Institute of NIH (TCGA RNA-seq data). No additional approval from the ethics committee was required. The present study complies with the requirements of data usage and publishing from NCBI GEO and TCGA databases.
Data preprocessing and DEGA
All microarray data were standardized by a normalized microarray preprocessing procedure using the affy package (version 1.60.0) in Bioconductor project (version 3.9.0, http://www.bioconductor.org/) (26), and RNA-seq data were subjected to normalization using the trimmed mean of M-values (TMM) method based on the edgeR package (version 3.26.3) in Bioconductor project (27).
DEG screening of microarray data was performed using the limma package (version 3.32.7) in Bioconductor project (28). The limma package employs the voom method, linear modeling and empirical Bayes moderation to assess DEGs, which yields more robust results, even with fewer microarrays. The edgeR package (version 3.26.3) in Bioconductor project was used to screen the DEGs between LUAD tissue and non-LUAD normal lung tissue samples of TCGA RNA-seq data (27).
WGCNA
Gene coexpression was analyzed using a WGCNA method, and WGCNA was performed using the WGCNA package (version 1.13) (29). First, an adjacency matrix was converted according to the gene expression matrix. Based on the adjacency matrix, the unsupervised coexpression relationship of each gene pair was computed by Pearson correlation coefficients. The soft threshold β was used to strengthen the correlation adjacency matrix, and the parameter β of each dataset was selected according to its scale-free topology criterion. Second, a topological matrix was converted according to the strengthened adjacency matrix, and the correlation of each gene pair was measured using the topological overlap measure (TOM). On the basis of TOM-based dissimilarity (1-TOM), the genes with coherent expression profiles were classified into a gene module using the average linkage hierarchical clustering method. Gene coexpression module was identified from the system cluster tree by a dynamic cutting algorithm. The modules with 75% similarity were merged into one module, and the representative gene in a module was identified as the module eigengene (ME). The correlation between the ME and gene module was defined as the module membership (MM). The gene differential expression was measured using the P value from t-test method between LUAD and normal lung tissues, and gene significance (GS) was computed by log10 transformation of the p value. The average GS was defined as the module significance (MS) of the module, and the MS indicated the correlation between the module and LUAD. A detailed description of the WGCNA method can be obtained from the original article (29).
PPI network construction and highly correlated module identification
The interactive relationships among DEGs encoding proteins were elucidated by constructing a PPI network, and the interactive relationships between gene pairs were retrieved from the online STRING database (version 11.0, https://string-db.org/) (30). Gene pairs with a combined score ≥0.7 were filtered to construct the PPI network, and the PPI network was established and visualized using Cytoscape software (version 3.7.0, http://www.cytoscape.org/) (31). Based on the topological properties of the whole PPI network, the highly correlated modules (subnetwork) were extracted from the whole PPI network using a Molecular COmplex DEtection (MCODE) algorithm. The MCODE analysis was performed using the plugin MCODE (version 1.5.1) in Cytoscape software (32). The threshold parameters were set as degree cut-off =4, node score cut-off =0.6, K-core =4 and max. depth =100.
Essential genes identification
Key genes were identified using seven centrality analyses in the PPI subnetwork (33). Seven centrality methods were Degree Centrality, Subgraph Centrality, Network Centrality, Eigenvector Centrality, Closeness Centrality, Betweenness Centrality and Information Centrality. The plugin CytoNCA (version 2.1.6) was used to perform the Centrality analyses in the Cytoscape software (34). The centrality score of each gene was computed by the centrality analyses, and the genes with higher centrality scores were identified as key genes. The intersecting genes of key genes were identified as the essential genes.
Identification of LUAD-specific prognostic gene signature
The LUAD patients from the TCGA database were used to perform the survival analysis. The Kaplan-Meier (KM) estimate and log-rank (LR) test were used to evaluate the associations between the essential genes and the OS of LUAD patients in the survival package (version 2.43-3, https://CRAN.R-project.org/package=survival). The group cut off was set to 50%, and the LR P value, hazard ratio (HR) and 95% confidence interval (CI) were computed. A P<0.05 indicated the statistical significant of the association between an essential gene and the OS of LUAD patients. A univariate Cox proportional hazards regression model was applied to evaluate the associations between the essential genes and the OS of LUAD patients. The same characteristic parameters obtained via the LR method were computed, and the same significant P value criterion was set. A multivariate Cox hazards regression model with the stepwise method was applied to assess the prognostic value for LUAD patients using the survival package in R project. The essential gene combination in the optimal Cox hazard regression model was used for further analyses. The hazards model was established as follows:
Risk score = ExpDEG1*CoeDEG1 + ExpDEG2*CoeDEG2+ ExpDEG3*CoeDEG3 …+ ExpDEGn*CoeDEGn
where “ExpDEGn” represented the expression level of DEGn and “CoeDEGn” denoted the regression coefficient from the multivariate Cox regression model (35). On the basis of the median of the above risk scores, LUAD patients were divided into the low-risk and high-risk groups. The survival ROC package (version 1.0.3) was used to construct the receiver operating characteristic (ROC) curve, and the ROC was used to measure the risk prediction rate of DEGs between the low- and high-risk groups.
LASSO Cox regression model analysis was performed using the glmnet package (version 4.0-2) based on R. The formula used to calculate the risk score is the same as that in the multivariate Cox hazards regression model, and the statistical methods are the same as those in the multivariate Cox hazards regression model.
Diagnostic analysis of prognostic genes
Diagnostic analysis of prognostic genes was performed using the support vector machine (SVM) method. The SVM classification model was constructed using the e1071 package (version 1.7-3) based on R. The radial basis function was applied in the SVM kernels, and the mRNA profiles were classified using 100 independent repetitions of 10-fold cross-validation. The specificity, sensitivity and accuracy were calculated.
Identification of candidate small-molecule drugs
Potential small-molecule drugs of the candidate prognostic genes were searched using three databases including CMap (https://portals.broadinstitute.org/cmap/) (36), L1000FWD (http://amp.pharm.mssm.edu/L1000FWD/) (37) and DGIdb (http://www.dgidb.org/) (38). The intersection of identified small molecules was indicative of potential candidate adjuvant drugs for use in LUAD patients.
Expression validation of essential genes by transcriptome sequencing data
To verify the expression of essential genes between LUAD tissue and normal lung tissue by bioinformatics methods, the transcriptomes of 10 nonsmoking LUAD patients of 35–50 years old (5 male and 5 female LUAD patients in early stages) from Xuanwei City (one of the areas with the highest morbidity and mortality of LC in China) in China were sequenced. All tissues were obtained from The First People’s Hospital of Yunnan Province. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of The First People’s Hospital of Yunnan Province (No. 2017YY227). Informed consent was taken from all the patients.
Transcriptome sequencing data were generated using the Illumina HiSeq2500 platform with the paired-end sequencing method by Beijing Novogene Technology Co., Ltd. DEGs between LUAD tissues and normal lung tissues samples were screened using the DESeq2 (version 1.24.0) package in Bioconductor project (39), and |logFC|>1 and FDR<0.01 were set as the cut-off criteria.
Statistical analysis
We performed data analysis according to the characteristics of each dataset on the basis of R software (version 3.6.3). For microarray datasets, DEGs were screened using the unpaired t-test in the limma package and a |Log2(fold change) (logFC)|>1 and a false discovery rate (FDR) <0.05 (P<0.05) were set as the cut-off criteria. For RNA-seq dataset, the quantile-adjusted conditional maximum likelihood (qCML) method in the edgeR package was used to identify DEGs, and the statistical significant of the difference was set for |logFC|>1.5 and FDR<0.01 on the basis of large-scale samples. The construction of prognostic signature was performed by univariate and multivariate COX regression analyses. Survival analysis was conducted by KM method and LR test, and P<0.05 indicated that the difference was statistically significant. Diagnostic analysis was performed using the SVM method. Gene coexpression analysis was performed using WGCNA, and gene significance was indexed by log10 transformation of the P value of the t-test measuring differential expression between LUAD and normal lung tissue samples. The expressions of key DEGs in prognostic and diagnostic signatures were analyzed using the paired-sample datasets including GSE7670 and GSE19804, and the paired t-test method was used to compare the expression difference between tumor samples and normal samples. A P<0.05 indicated that the difference was statistically significant in two groups.
Results
DEGs identification and functional analysis
To identify DEGs involved in LUAD, DEGA was performed. According to |logFC|>1 and FDR <0.05, totals of 623 (Dataset A, 189 upregulated and 434 downregulated), 1,387 (Dataset B, 424 upregulated and 963 downregulated), 1,343 (Dataset C, 492 upregulated and 851 downregulated) DEGs were identified. According to |logFC|>1.5 and FDR <0.01, 11,450 (8,940 upregulated and 2,510 downregulated) DEGs were identified. Overlapping analysis showed that 235 common DEGs (74 upregulated and 161 downregulated) were identified (available online: https://cdn.amegroups.cn/static/public/tcr-21-526-1.pdf).
To better understand the roles of 235 common DEGs in LUAD, functional enrichment analyses including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Reactome pathways (RP) and Disease Ontology (DO) analyses were performed using the online STRING database (version 11.0) and clusterProfiler package (version 3.12.0) in Bioconductor project (30,40). GO analysis showed that 235 dysregulated genes were separately significantly enriched in 163 upregulated and 537 downregulated biological processes (BPs) (P<0.05), 18 upregulated and 29 downregulated molecular functions (MFs) (P<0.05), and 22 upregulated and 36 downregulated cellular components (CCs) (P<0.05). The top 5 GO terms with the most significant P values are shown in Table 1. Among the enriched BPs, upregulated BPs were mainly related to the cell cycle process, and downregulated BPs were mainly associated with the biological regulation (Table 1). KEGG pathway analysis showed that 2 upregulated pathways including the cell cycle (P=0.00062) and ECM-receptor interaction (P=0.0101) were significantly enriched (Table 1), and no significantly downregulated pathways were enriched. RP analysis showed that 235 dysregulated genes were separately enriched in 1 upregulated and 1 downregulated pathways (Table 1), and the 2 RPs were upregulated collagen degradation (P=0.0068) and downregulated hemostasis (P=0.0045), respectively. DO analysis showed that up- and down-regulated DEGs were separately associated with 32 and 9 diseases, and the top 5 DO terms are listed in Table 1. All upregulated DO terms were significantly associated with many types of cancers, and the most significantly upregulated DO term was non-small cell lung carcinoma (P=0.000764424) (Table 1).
Table 1
| Term ID | Term description | Observed | FDR |
|---|---|---|---|
| Biological process | |||
| Up-regulated | |||
| GO:1903047 | Mitotic cell cycle process | 21 | 5.01E-12 |
| GO:0022402 | Cell cycle process | 23 | 1.11E-10 |
| GO:0051301 | Cell division | 14 | 1.91E-06 |
| GO:0000280 | Nuclear division | 11 | 2.81E-06 |
| GO:0007059 | Chromosome segregation | 10 | 1.51E-05 |
| Down-regulated | |||
| GO:0051239 | Regulation of multicellular organismal process | 67 | 1.36E-13 |
| GO:0009653 | Anatomical structure morphogenesis | 51 | 1.80E-10 |
| GO:0050793 | Regulation of developmental process | 56 | 2.61E-10 |
| GO:0072359 | Circulatory system development | 32 | 2.61E-10 |
| GO:0001944 | Vasculature development | 25 | 4.38E-10 |
| Molecular function | |||
| Up-regulated | |||
| GO:0005515 | Protein binding | 45 | 0.00063 |
| GO:0042802 | Identical protein binding | 20 | 0.00099 |
| GO:0042803 | Protein homodimerization activity | 13 | 0.0016 |
| GO:0046983 | Protein dimerization activity | 16 | 0.0021 |
| GO:0005488 | Binding | 61 | 0.0034 |
| Down-regulated | |||
| GO:0005539 | Glycosaminoglycan binding | 12 | 0.00024 |
| GO:0005102 | Signaling receptor binding | 31 | 0.00048 |
| GO:0005488 | Binding | 126 | 0.00048 |
| GO:0005515 | Protein binding | 83 | 0.00048 |
| GO:0017046 | Peptide hormone binding | 6 | 0.00048 |
| Cellular component | |||
| Up-regulated | |||
| GO:0005576 | Extracellular region | 28 | 1.14E-05 |
| GO:0005819 | Spindle | 10 | 5.09E-05 |
| GO:0000776 | Kinetochore | 6 | 0.00092 |
| GO:0000940 | Condensed chromosome outer | 3 | 0.001 |
| GO:0005615 | Extracellular space | 15 | 0.001 |
| Down-regulated | |||
| GO:0005576 | Extracellular region | 55 | 1.28E-09 |
| GO:0005615 | Extracellular space | 31 | 7.89E-07 |
| GO:0005886 | Plasma membrane | 74 | 9.46E-06 |
| GO:0009986 | Cell surface | 22 | 9.46E-06 |
| GO:0031226 | Intrinsic component of plasma membrane | 34 | 3.13E-05 |
| KEGG pathway | |||
| Up-regulated | |||
| hsa04110 | Cell cycle | 6 | 0.00062 |
| hsa04512 | ECM-receptor interaction | 4 | 0.0101 |
| Reactome pathway | |||
| Up-regulated | |||
| HSA-1442490 | Collagen degradation | 3 | 0.0068 |
| Down-regulated | |||
| HSA-109582 | Hemostasis | 15 | 0.0045 |
| Disease ontology | |||
| Up-regulated | |||
| DO:3908 | Non-small cell lung carcinoma | 15 | 0.0007644 |
| DO:3459 | Breast carcinoma | 13 | 0.0027271 |
| DO:0050904 | Salivary gland carcinoma | 5 | 0.0088499 |
| DO:8850 | Salivary gland cancer | 5 | 0.0088499 |
| DO:0060084 | Cell type benign neoplasm | 13 | 0.0088499 |
| Down-regulated | |||
| DO:6000 | Congestive heart failure | 14 | 0.0134019 |
| DO:6432 | Pulmonary hypertension | 8 | 0.0134019 |
| DO:5844 | Myocardial infarction | 15 | 0.0134019 |
| DO:3393 | Coronary artery disease | 17 | 0.0134019 |
| DO:1936 | Atherosclerosis | 16 | 0.0261586 |
DEG, differentially expressed gene; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix.
Interactive relationships among 235 DEGs and essential gene identification
To elucidate the interactive relationships among 235 DEGs, a PPI network was constructed. At a minimum required interaction score = high confidence 0.7, a total of 108 DEGs was filtered into the PPI network, and a PPI network with 108 nodes and 320 edges was established (Figure 2A). Three highly correlated modules were identified in the whole PPI network, and the module with the highest score (score =19.789) included 20 nodes and 188 edges (Figure 2B). The 20 genes had high centrality scores (Table 2) and were identified as essential genes including UBE2C, ZWINT, MELK, CCNB2, RRM2, CEP55, BIRC5, TTK, KIF20A, CDKN3, TOP2A, NUSAP1, ASPM, CDC20, BUB1B, CCNB1, TYMS, MCM4, CENPF and KIF4A. These genes were mainly associated with many types of cancers including NSCLC and LUAD (Figure 2C) and were mainly enriched within pathways related to the cell cycle (Figure 2D). All the genes were highly expressed in LUAD tissue (all P<0.001) (Figure 2E), and every pair of genes showed a strong positive correlation in expression (all P<0.001) (Figure 2F).
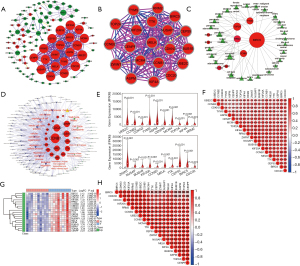
Table 2
| Rank | Gene | Subgraph | Eigenvector | Information | LAC | Betweenness | Closeness | Network | Degree |
|---|---|---|---|---|---|---|---|---|---|
| 1 | UBE2C | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 2 | ZWINT | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 3 | MELK | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 4 | CCNB2 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 5 | RRM2 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 6 | CEP55 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 7 | BIRC5 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 8 | TTK | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 9 | KIF20A | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 10 | CDKN3 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 11 | TOP2A | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 12 | NUSAP1 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 13 | ASPM | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 14 | CDC20 | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 15 | BUB1B | 7504268.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 16 | CCNB1 | 7504266.50 | 0.226 | 10.47 | 17.79 | 0.222 | 1.0 | 19.00 | 19 |
| 17 | TYMS | 6799982.00 | 0.215 | 10.22 | 16.89 | 0.111 | 0.95 | 17.88 | 18 |
| 18 | MCM4 | 6799982.00 | 0.215 | 10.22 | 16.89 | 0.111 | 0.95 | 17.88 | 18 |
| 19 | CENPF | 6799982.00 | 0.215 | 10.22 | 16.89 | 0.111 | 0.95 | 17.88 | 18 |
| 20 | KIF4A | 6799982.00 | 0.215 | 10.22 | 16.89 | 0.111 | 0.95 | 17.88 | 18 |
LAC, local average connectivity.
Expression validation of 20 essential genes
To validate the differential expressions of 20 essential genes between LUAD and normal lung tissues, 20 transcriptomes from 10 nonsmoking LUAD patients in an early stage were analyzed. According to |logFC|>1 and FDR<0.01, 2,360 DEGs (1,302 upregulated and 1,058 downregulated) were identified (available online: https://cdn.amegroups.cn/static/public/tcr-21-526-2.pdf). Except TYMS, 19 DEGs were consistent with the results from integrative data, and the expressions of 19 DEGs were visualized using the heatmap shown in Figure 2G. Similarly, there were stronger positive correlations in expression among these genes (all R2>0.55, most P<0.001) (Figure 2H). The RNA-seq data have been deposited in the NCBI Short Read Archive (Accession number: PRJNA561283). Given the high consistency of the identified DEGs, the 20 DEGs were selected for further analyses.
Consensus clustering of 20 essential genes and relationships with distinct clinical outcomes and clinicopathological features
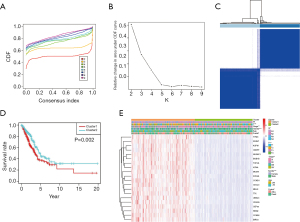
To investigate the relationships between 20 essential genes and distinct clinical outcomes and clinicopathological features, consensus clustering was performed. On the basis of the expression similarity of 20 essential genes, k=2 was selected according to clustering stabilities increasing from k=2 to 9 in the TCGA dataset (Figure 3A,B) and clustered into two subgroups (Figure 3C). The two subgroups were significantly related to the prognosis of LUAD patients, and the cluster2 subgroup had a higher survival rate (P=0.002, Figure 3D). Clinicopathological features were compared between the two subgroups. The results showed that all these genes were lowly expressed in the cluster2 subgroup, and the cluster2 subgroup was significantly correlated with an earlier N stage, T stage and pathological grade (P<0.05, 0.001 and 0.01, separately), as well as with fewer female and dead patients (P<0.001 and 0.05, separately) (Figure 3E). The results explained why patients had a higher OS in the cluster2 subgroup.
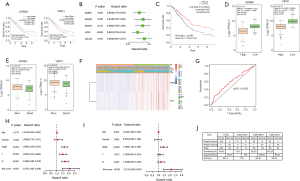
Survival analysis and prognostic model of 20 essential genes
To elucidate the relationships between 20 essential genes and the OS of LUAD patients, survival analysis was performed. Univariate Cox regression showed that all 20 essential genes were significantly correlated with the OS of LUAD patients (all P<0.01) and were risky genes with HR >1 (Figure 4A). To better predict the clinical outcomes of LUAD with 20 essential genes, the LASSO Cox regression algorithm was applied and 12 essential genes (ZWINT, MELK, CCNB2, RRM2, TTK, KIF20A, TOP2A, ASPM, CDC20, CCNB1, TYMS and KIF4A) were selected to build the risk signature based on the minimum criteria (Figure 4B,C). The risk score of each patient was calculated, and LUAD patients were divided into high-risk and low-risk subgroups on the basis of the median risk score. A significant difference in OS was observed between the two subgroups (P=1e-05), and the low-risk group showed a higher survival rate (Figure 4D). The KM estimate and LR test showed that 12 genes were significantly associated with the OS of LUAD patients (all P<0.05, Figure S1).
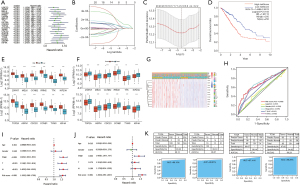
The expression levels of 12 genes were analyzed between the high-risk and low-risk groups. The results showed that all the genes were significantly highly expressed in high risk group (all P<0.001, Figure 4E). The expression levels of 12 genes were also compared between alive and dead patient groups. The result showed that 12 genes were significantly lowly expressed in the alive patient group (all P<0.01, separately, Figure 4F), which indicates that low expressions of these genes contribute to a low risk of LUAD patients and lengthen survival of LUAD patients.
To elucidate the associations between risk scores and clinicopathological features, clinicopathological features were analyzed between the high-risk and low-risk subgroups. We observed significant differences between the two risk subgroups with respect to the N stage (P<0.01), T stage (P<0.01), pathological stage (P<0.001) and survival status (P<0.001) (Figure 4G). The ROC curve showed that the risk score and pathological stage could better predict the three-year OS for LUAD patients (AUC =0.686 and 0.675, separately) (Figure 4H).
To determine whether the risk signature is an independent prognostic indicator, univariate and multivariate Cox regression analyses were performed. By univariate Cox regression analysis, the risk score, N stage, T stage and pathological stage were significantly correlated with the OS of LUAD patients (all P<0.001) (Figure 4I). Multivariate Cox regression analysis showed that risk score and pathological stage were significantly correlated with the OS of LUAD patients (P<0.001 and =0.003, separately) (Figure 4J). These results indicate that the risk score can independently predict the OS in LUAD patients.
Diagnostic model based on 12 essential genes related to prognosis
To evaluate diagnostic power of 12 essential genes related to the prognosis of LUAD patients, a diagnostic model of 12 genes was constructed using the SVM classification model. The results showed that both the specificity and sensitivity of classification exceed 93% in the three datasets (Figure 4K). The AUCs were over 95% in the three datasets (Figure 4K), which indicates that 12 genes are very effective as diagnostic biomarkers in predicting the diagnosis of LUAD patients.
Coexpression network construction and module analysis
To better understand the gene coexpression relationships in different tissue types, WGCNA was performed. By normalization, 12,410 (Dataset A), 20,217 (Dataset B), 17,702 (Dataset C) and 31,343 (Dataset D) genes were selected to construct the gene coexpression network. On the basis of a scale-free topology criterion R2>0.8, power β=7 (Dataset A, R2=0.84), 18 (Dataset B, R2=0.85), 4 (Dataset C, R2=0.85) and 2 (Dataset D, R2=0.86) were selected as soft thresholds to convert the Pearson correlation matrix into a strengthened adjacency matrix, separately (Figure 5A, Figure S2A). The TOM of each gene pair was calculated, and 20, 17, 35 and 36 coexpression modules were separately identified by average linkage hierarchical clustering according to a TOM-based dissimilarity measure (1-TOM) in four datasets (Figure 5B, Figure S2B). The correlation analysis between ME and LUAD showed that the brown (including 1,875 genes, cor=−0.87 and P=6.20E-50), dark turquoise (including 1,895 genes, cor=−0.83 and P=1.45E-31), yellow (including 910 genes, cor=−0.94 and P=5.37E-15) and blue (including 3,025 genes, cor=−0.88 and P=1.9E-190) modules were separately the most significant modules in the four datasets (Figure 5C, Figure S2C). The correlation analysis of the MMs in these modules showed that these MMs had the most significant correlation with LUAD (Dataset A, cor=0.87 and P<1e-200; Dataset B, cor=0.77 and P<1e-200; Dataset C, cor=0.95 and P<1e-200; Dataset D, cor=0.98 and P<1e-200; Figure 5D and Figure S2D). GSs across modules showed that these modules had the highest GS values (Figure 5E, Figure S2E). The four modules were selected as key coexpression gene modules for further analyses.
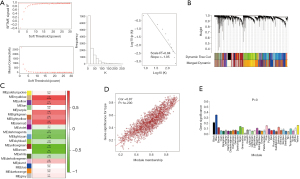
Key gene identification in the four most significant modules
To identify the key genes among the four most significant modules, DEGA and overlapping analyses were performed in the four modules. The results showed that 392 (Dataset A), 869 (Dataset B), 483 (Dataset C) and 2,270 (Dataset D) DEGs were separately identified (Figure 6A), and 52 common DEGs (5 upregulated and 47 downregulated) were identified (Figure 6B, Table 3). These DEGs were mainly involved in blood vessel development (GO:0001568, P=1.80e-6), vasculature development and regulation (GO:1901342, P=2.17e-6) and tube development (GO:0035295, P=6.03e-6). The expressions of 52 common DEGs are shown using a heatmap in Figure 6C.
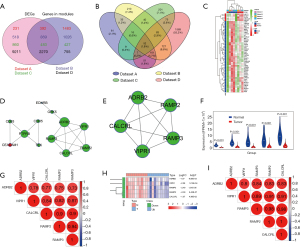
Table 3
| Gene symbol | Dataset A | Dataset B | Dataset C | Dataset D | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| logFC | FDR | logFC | FDR | logFC | FDR | logFC | FDR | ||||
| Up-regulated | |||||||||||
| PYCR1 | 1.0834 | 2.60E-34 | 1.2966 | 2.01E-26 | 2.7821 | 4.05E-07 | 3.7387 | 9.68E-93 | |||
| CEACAM1 | 1.1290 | 7.28E-18 | 1.2744 | 7.37E-11 | 1.2071 | 1.90E-02 | 2.0955 | 2.63E-30 | |||
| ABCC3 | 1.3676 | 3.89E-22 | 1.9165 | 1.23E-19 | 3.7939 | 7.01E-09 | 2.5686 | 1.13E-37 | |||
| GOLM1 | 1.8824 | 7.93E-36 | 2.0276 | 8.61E-29 | 1.8870 | 4.45E-07 | 2.6754 | 4.03E-65 | |||
| HMGB3 | 2.1441 | 1.80E-36 | 1.7898 | 3.38E-13 | 1.9850 | 5.23E-04 | 3.5250 | 6.97E-48 | |||
| Down-regulated | |||||||||||
| SFTPC | 3.7846 | 8.10E-26 | 3.1933 | 6.72E-11 | 2.7964 | 5.01E-04 | 4.7077 | 9.03E-69 | |||
| FCN3 | 3.6158 | 1.00E-46 | 3.4480 | 4.02E-20 | 3.8986 | 9.14E-06 | 4.4931 | 3.91E-172 | |||
| TMEM100 | 3.5293 | 1.35E-41 | 3.6015 | 7.46E-18 | 3.9073 | 6.85E-09 | 4.3314 | 6.7E-141 | |||
| ABCA8 | 2.9376 | 7.81E-50 | 2.8246 | 2.07E-16 | 1.3704 | 2.67E-05 | 2.8318 | 1.23E-71 | |||
| CDH5 | 2.5828 | 1.01E-51 | 2.2366 | 7.80E-24 | 2.2076 | 2.85E-08 | 2.5234 | 4.06E-148 | |||
| GPM6A | 2.5467 | 3.4E-47 | 3.9236 | 2.82E-28 | 2.5505 | 3.51E-08 | 4.4338 | 9.26E-122 | |||
| EDNRB | 2.5449 | 9.55E-50 | 2.9911 | 4.45E-22 | 2.6127 | 6.85E-09 | 3.4758 | 1.75E-177 | |||
| GRK5 | 2.4935 | 2.23E-57 | 1.8852 | 2.82E-13 | 2.0877 | 2.73E-08 | 2.4636 | 2.62E-128 | |||
| TEK | 2.4787 | 1.72E-52 | 2.6268 | 3.02E-24 | 2.6799 | 2.85E-08 | 3.2279 | 6.82E-191 | |||
| CLDN18 | 2.4480 | 9.25E-32 | 3.2886 | 9.02E-14 | 2.2100 | 1.33E-03 | 3.5131 | 2.38E-38 | |||
| CA4 | 2.3105 | 9.1E-46 | 3.2181 | 2.02E-31 | 3.5226 | 3.35E-08 | 4.3314 | 2.42E-104 | |||
| TGFBR3 | 2.2710 | 4.24E-42 | 2.1643 | 1.94E-16 | 2.0791 | 1.06E-08 | 2.5536 | 6.25E-89 | |||
| LDB2 | 2.2329 | 2.04E-45 | 2.1822 | 3.70E-21 | 1.9013 | 1.88E-09 | 2.5873 | 8.98E-170 | |||
| CAV2 | 2.1706 | 1.36E-21 | 2.1045 | 2.90E-12 | 1.7610 | 1.53E-07 | 2.3153 | 1.51E-70 | |||
| EMP2 | 2.0621 | 3.99E-47 | 1.8399 | 9.91E-18 | 1.6546 | 7.72E-07 | 2.7098 | 4.00E-179 | |||
| VIPR1 | 2.0389 | 4.11E-37 | 1.9417 | 2.10E-15 | 2.6302 | 7.84E-10 | 3.2094 | 3.43E-127 | |||
| RAMP3 | 2.0118 | 1.71E-38 | 2.0534 | 1.66E-21 | 2.8286 | 7.72E-08 | 3.1995 | 5.78E-185 | |||
| GIMAP6 | 1.9154 | 8.16E-34 | 1.9864 | 1.40E-17 | 1.7776 | 1.18E-06 | 2.0685 | 2.62E-91 | |||
| DOCK4 | 1.8372 | 1.05E-30 | 1.4940 | 3.12E-20 | 1.2694 | 5.15E-07 | 1.5397 | 1.59E-59 | |||
| RAMP2 | 1.8250 | 1.01E-45 | 1.7628 | 2.60E-18 | 2.6198 | 2.92E-07 | 2.8586 | 1.83E-166 | |||
| RASIP1 | 1.8037 | 2.46E-45 | 2.0014 | 1.30E-28 | 2.1561 | 2.23E-08 | 2.2819 | 2.09E-101 | |||
| EMCN | 1.7935 | 1.17E-42 | 2.5049 | 3.43E-17 | 1.7784 | 7.21E-05 | 2.7228 | 1.43E-114 | |||
| SLC6A4 | 1.7909 | 1.87E-26 | 2.9093 | 1.14E-17 | 4.5929 | 7.01E-09 | 6.1370 | 9.64E-174 | |||
| IL33 | 1.7537 | 8.30E-29 | 2.0729 | 1.04E-12 | 1.5444 | 3.11E-05 | 2.0778 | 4.94E-41 | |||
| ERG | 1.7476 | 5.85E-40 | 1.4934 | 9.67E-14 | 2.0968 | 1.26E-05 | 2.1248 | 3.21E-115 | |||
| ADRB2 | 1.6150 | 2.42E-38 | 1.9791 | 6.43E-20 | 1.4662 | 1.3E-04 | 3.0541 | 1.69E-128 | |||
| ANXA3 | 1.5754 | 1.37E-17 | 1.7950 | 1.01E-10 | 1.4992 | 2.27E-04 | 1.8378 | 2.18E-41 | |||
| ITM2A | 1.5484 | 3.02E-32 | 1.6888 | 3.30E-15 | 1.3892 | 2.10E-07 | 2.0161 | 1.33E-67 | |||
| TBX3 | 1.4951 | 4.79E-31 | 1.3848 | 1.96E-13 | 1.9755 | 3.53E-05 | 2.1108 | 1.26E-71 | |||
| JAM2 | 1.4844 | 7.69E-50 | 1.9511 | 8.55E-20 | 1.8270 | 7.72E-08 | 2.4928 | 1.57E-150 | |||
| CD93 | 1.4564 | 1.46E-28 | 1.6924 | 7.67E-17 | 1.8429 | 6.54E-07 | 2.1842 | 5.72E-103 | |||
| GHR | 1.4503 | 5.31E-33 | 1.8830 | 2.24E-15 | 1.5769 | 5.72E-04 | 2.0160 | 2.21E-45 | |||
| STXBP6 | 1.4350 | 1.56E-46 | 2.6371 | 1.02E-19 | 2.9172 | 2.99E-11 | 3.3623 | 3.84E-103 | |||
| TMEM47 | 1.4317 | 2.40E-18 | 1.4365 | 1.25E-12 | 1.3715 | 1.49E-04 | 1.7287 | 2.81E-55 | |||
| FERMT2 | 1.3605 | 4.41E-31 | 1.2932 | 7.35E-08 | 1.0452 | 5.26E-05 | 1.3922 | 9.59E-63 | |||
| SEMA6A | 1.3278 | 9.00E-21 | 1.6010 | 4.60E-12 | 2.3603 | 1.33E-10 | 2.5439 | 1.49E-95 | |||
| SEMA5A | 1.3200 | 1.03E-30 | 1.5949 | 8.43E-19 | 1.9359 | 2.09E-05 | 2.5148 | 6.65E-77 | |||
| PTPRB | 1.3143 | 3.05E-36 | 2.1648 | 7.02E-21 | 1.9385 | 1.05E-05 | 2.5855 | 1.33E-119 | |||
| CALCRL | 1.3001 | 2.6E-22 | 2.0359 | 2.50E-09 | 2.2940 | 1.28E-04 | 2.7443 | 8.75E-150 | |||
| SASH1 | 1.2749 | 2.18E-41 | 1.4206 | 4.38E-18 | 1.6155 | 2.34E-08 | 1.8352 | 2.13E-102 | |||
| S1PR1 | 1.2691 | 1.09E-40 | 2.0206 | 1.38E-22 | 2.0551 | 8.06E-06 | 2.8252 | 4.29E-189 | |||
| LMCD1 | 1.2579 | 5.45E-29 | 1.1033 | 1.87E-13 | 1.4161 | 3.59E-04 | 1.7207 | 8.72E-84 | |||
| SPTBN1 | 1.2511 | 6.33E-24 | 1.6536 | 2.06E-09 | 1.5544 | 6.94E-06 | 1.7150 | 1.41E-95 | |||
| EML1 | 1.2483 | 1.09E-35 | 1.4229 | 2.21E-12 | 1.3991 | 4.63E-06 | 1.6725 | 2.98E-51 | |||
| PODXL | 1.1869 | 1.80E-26 | 1.1076 | 3.11E-15 | 1.2530 | 7.96E-05 | 1.0640 | 3.71E-28 | |||
| KL | 1.1122 | 5.76E-30 | 2.2987 | 2.82E-24 | 2.0117 | 3.83E-05 | 2.3051 | 2.19E-41 | |||
| PDK4 | 1.1117 | 3.07E-18 | 1.8544 | 1.058E-06 | 1.8622 | 4.63E-04 | 2.3871 | 2.32E-53 | |||
DEG, differentially expressed gene; FC, fold change; FDR, false discovery rate.
PPI network construction and essential gene identification based on 52 common DEGs
To elucidate the interactive relationships among 52 DEGs, a PPI network was constructed. At a minimum required interaction score = high confidence 0.7, a total of 12 (1 upregulated and 11 downregulated) among 52 DEGs was filtered into the PPI network. A PPI network with 12 nodes and 18 edges was established (Figure 6D). One highly correlated module was identified in the whole PPI network, and the module included 5 nodes and 10 edges (score =5.00) (Figure 6E). Centrality analysis showed that the 5 downregulated genes including ADRB2, RAMP2, CALCRL, VIPR1 and RAMP3 had the same centrality score (Table 4) and were identified as essential genes involved in LUAD. All the essential genes were lowly expressed in LUAD tissue (all P<0.001) (Figure 6F), and every pair of genes showed a strong positive correlation in expression (all R2>0.75 and P<0.001) (Figure 6G). On the basis of the data of 20 transcriptomes, the expressions of 5 genes were significantly downregulated in LUAD tissue (all P<0.01) (Figure 6H) and had strong correlations in expression (all R2>0.80 and P<0.001) (Figure 6I).
Table 4
| Rank | Gene | Subgraph | Degree | Eigenvector | Information | Betweenness | Closeness | Network |
|---|---|---|---|---|---|---|---|---|
| 1 | ADRB2 | 11.2139 | 4 | 0.4472 | 3.125 | 0 | 1.0 | 4.0 |
| 2 | VIPR1 | 11.2139 | 4 | 0.4472 | 3.125 | 0 | 1.0 | 4.0 |
| 3 | RAMP3 | 11.2139 | 4 | 0.4472 | 3.125 | 0 | 1.0 | 4.0 |
| 4 | RAMP2 | 11.2139 | 4 | 0.4472 | 3.125 | 0 | 1.0 | 4.0 |
| 5 | CALCRL | 11.2139 | 4 | 0.4472 | 3.125 | 0 | 1.0 | 4.0 |
Prognostic analysis based on 5 essential genes
To elucidate the relationships between 5 essential genes and the OS of LUAD patients, survival analysis was performed using the KM estimate and LR test. According to the P<0.05 cut-off criterion, ADRB2 (P=0.01009) and VIPR1 (P=0.01613) were identified as associated with the OS of LUAD patients (Figure 7A), and ADRB2 (HR =0.68215, 95% CI: 0.5081–0.9143) and VIPR1 (HR =0.70189, 95% CI: 0.5199–0.9369) were protective genes with HR <1 (Figure 7A). Higher expressions of the two genes resulted in a higher OS rate of LUAD patients (Figure 7A). Similar results were found in the univariate Cox regression analysis, and ADRB2 and VIPR1 had significant prognostic value in LUAD patients (Figure 7B). Higher mRNA expressions of ADRB2 (P=0.002, HR =0.854, 95% CI: 0.775–0.943) and VIPR1 (P<0.001, HR =0.831, 95% CI: 0.755–0.914) had lower HRs and resulted in a higher OS rate (Figure 7B). The multivariate Cox regression model with the stepwise method based on the 5 essential genes showed that ADRB2 and VIPR1 had significant prognostic value for the OS of LUAD patients (P=0.000124). A two-gene prognostic model was established, and the risk score of each patient was calculated. On the basis of the median of the risk scores, the LUAD patients were divided into high-risk and low-risk subgroups. The mortality rate of the high-risk subgroup was significantly higher than that of the low-risk group [43.24% (109 in 252 patients) vs. 29.37% (74 in 252 patients), P=0.001637], and the high-risk subgroup had a worse prognosis compared to the low-risk group (P=0.00225, HR =1.57692, 95% CI: 1.1749–2.212, Figure 7C).
The expression levels of ADRB2 and VIPR1 were analyzed between the high-risk and low-risk groups. The results showed that both ADRB2 and VIPR1 were significantly highly expressed in the low-risk group (both P<2.2e-16, Figure 7D). Further, the expression levels of ADRB2 and VIPR1 were observed between the alive and dead patient groups. The result showed that both ADRB2 and VIPR1 were significantly highly expressed in the alive patient group (P=0.001724 and 0.000709, separately, Figure 7E), which indicates that high expressions of ADRB2 and VIPR1 contribute to a low risk for LUAD patients and lengthen survival of LUAD patients.
To elucidate the associations between the risk scores and clinicopathological features, clinicopathological features were analyzed between the high- and low-risk subgroups. We observed significant differences between the two risk subgroups with respect to the T stage (P<0.01), gender (P<0.05) and survival status (P<0.05) (Figure 7F). The ROC indicates that the risk score can predict the prognosis of LUAD patients (Figure 7G).
To determine whether the risk signature is an independent prognostic indicator, univariate and multivariate Cox regression analyses were performed. According to univariate Cox regression analysis, the risk score, N stage, T stage and pathological stage were significantly correlated with the OS of LUAD patients (all P<0.001) (Figure 7H). Multivariate Cox regression analysis showed that risk score and pathological stage were significantly correlated with the OS of LUAD patients (both P<0.001) (Figure 7I). These results indicate that the risk score can independently predict the OS in LUAD patients.
Diagnostic analysis of the 2-gene signature
Through the diagnostic analysis of four datasets, the results showed that both the specificity and sensitivity of classification exceed 93% in the three datasets (Figure 7J), and the AUCs were over 94% (Figure 7J). Among these values, the AUC was 100% in the GSE102511 dataset, which indicates that the 2-gene signature has very high effectiveness as a diagnostic biomarker in predicting the diagnosis of LUAD patients.
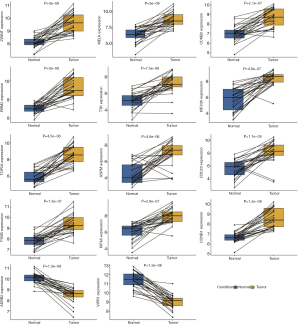
Expression analysis of 12-gene and 2-gene signatures in paired sample datasets
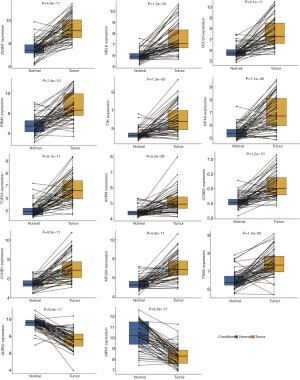
In this study, some paired samples were included in GSE7670 and GSE19804. To validate the expression of DEGs in 12-gene and 2-gene signatures, paired samples were selected and analyzed the expression difference between tumor and normal lung tissues using the paired t-test method. The results showed that 14 genes had statistical significant in expression between two groups in two datasets (all P<0.05, Figure 8 and Figure 9), which was consistent with the results obtained from the unpaired t-test method.
Identification of small-molecule drugs
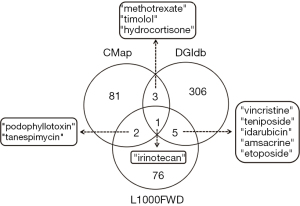
To identify potential adjuvant drugs based on 14 prognostic genes (12-gene and 2-gene signatures) to guide the therapy of LUAD patients, small-molecule drugs were screened using three databases including CMap, L1000FWD and DGIdb. Totals of 87, 84 and 315 small molecules were separately identified in three small-molecule databases, and 1 small molecule with the drug name irinotecan was simultaneously found (Figure 10). By pairwise comparison, 3 (podophyllotoxin, tanespimycin, and irinotecan), 4 (methotrexate, irinotecan, timolol, and hydrocortisone) and 6 (vincristine, teniposide, idarubicin, amsacrine, irinotecan, and etoposide) small molecules were found (Figure 10).
Discussion
LUAD, as the most commonly pathological subtype of LC and that accounts for more than 40% of LCs (3,4), is one of the leading causes of LC-related deaths (1). Despite recent advances in detection technologies and treatment methods, the 5-year OS rate of LUAD patients in all stages remains very poor, at less than 20% (3,6). The identification of diagnostic and prognostic biomarkers may contribute to improving the survival rate of LUAD patients. However, due to the high heterogeneity of LUAD, the results reported from different studies vary enormously, and some identified biomarkers are not widely accepted for predicting clinical outcomes. To identify consistently acceptable diagnostic and prognostic biomarkers to improve clinical outcomes, systematic analysis is essential to explore the molecular mechanisms involved in LUAD and identify some key diagnostic and prognostic signatures by integrating LUAD-related gene expression data. In this study, we systematically integrated five LUAD-related gene expression datasets using bioinformatics methods including WGCNA, DEGA, PPI network, and prognostic and diagnostic analyses to identify transcriptome characterization to mine the key genes involved in LUAD. Finally, 12-gene (ZWINT, MELK, CCNB2, RRM2, TTK, KIF20A, TOP2A, ASPM, CDC20, CCNB1, TYMS and KIF4A) and 2-gene (ADRB2 and VIPR1) signatures were identified in LUAD and may serve as diagnostic and prognostic markers of LUAD patients. Furthermore, eleven small-molecule drugs related to the 12-gene and 2-gene signatures were identified, providing novel insights for LUAD therapeutic studies.
As can be seen from the “Methods” and “Results” sections, 12-gene and 2-gene signatures were identified by a systematical analysis based on DEGs obtained from an unpaired t-test method. Before discussing the potential applicability of two signatures as diagnostic and prognostic markers, we must eliminate several doubts that readers may have on the processing method: (I) GSE7670 and GSE19804 datasets included many paired samples, why used an unpaired t-test for DEGA? (II) Why were GSE7670 and GSE10072 datasets merged into one dataset? (III) Why the microarray datasets and RNA-seq dataset do not use the same cutoff standard when performing DEGA? For the first question, theoretically it is more suitable to use paired t-test to process paired samples. We tried to use the paired t-test method to screen DEGs on paired sample datasets. However, few DEGs with |logFC|>1 and adjusted P<0.05 were identified using the method. Especially, the adjusted P values were significantly improved in the paired t-test model. We consider that the adjusted P values may be over-corrected in the test model when the sample number is smaller. The main reason may be that the degree of freedom is reduced and the gene expression matrix is re-standardized after samples were paired. It is not rigorous to only rely on logFC value to identify differentially expressed genes. In order to compensate the deficiency, we used the paired t-test method to analyze the expression differences of DGEs in the two signatures based on single gene. The results showed that all DEGs had statistical significance in the expression in GSE7670 and GSE19804 datasets (P<0.05, Figure 8 and Figure 9). For the second question, the sample number is more and the statistical reliability of the data is stronger in theory. Due to the same microarray platform, two datasets were merged into one dataset by reconstructing gene expression profile after background was corrected using a normalized microarray preprocessing procedure in the affy package. To a greater extent, the method eliminates some technical errors than the method of simply merging gene expression profiles. This method is expected to improve statistical power to get more reliable results. Judging from the results of this study, the method is feasible. For the third question, RNA-seq dataset included more samples and genes than microarray dataset, which indicates that RNA-seq dataset can have a better “resolution” in identifying DEGs. So, a more stringent cutoff standard was used for RNA-seq dataset.
In the 12-gene signature, all the genes were enriched in biological pathways related to the cell cycle (Figure 2C). As we all know, the cell cycle is one of the most critical pathways closely associated with cancers, and an abnormal cell cycle will initiate malignant tumors (41,42). So far, some genes related to the cell cycle have also been confirmed to play important roles in LC and serve as potential prognostic candidates to predict the survival of NSCLC patients, such as ZWINT and CDC20, as well as BUB1B (43,44). The current results were consistent with a previous study (44). ZWINT is an important regulatory protein and plays key roles in chromosome movement and mitotic checkpoints (45,46). Some studies showed that the dysfunction of ZWINT resulted in many types of cancers such as breast and ovarian cancers (45), and the overexpression of ZWINT predicted a poor prognosis (47,48). However, a few studies also showed that ZWINT was a protective gene in hepatocellular carcinoma, and its increased expression contributed to a good prognosis (49,50). In this study, ZWINT was identified as a risky factor in LUAD, and its upregulated expression resulted in a poor prognosis. This result was consistent with previous studies indicating that a high expression of ZWINT was closely related to a poor prognosis of LUAD patients (45). CCNB2, which is a member of the cyclin family, is one of the essential components of the cell cycle regulatory machinery and plays a key role in transforming growth factor beta-mediated cell cycle control. CCNB2 has been reported to associate with many cancers (51), and its overexpression predicted a poor prognosis in NSCLC patients (52). CCNB1 is another member of the cyclin family, and its gene product plays critical role in controlling the G2/M transition phase of the cell cycle by forming the maturation-promoting factor (MPF) with CDC2 (53). The upregulation of CCNB1 in tumor tissue predicted a worse prognosis in hepatocellular carcinoma patients (54), and it may serve as potential therapeutic target (55). Presently, a few studies have shown that CCNB1 plays a role in NSCLC (56), and its polymorphisms were associated with clinical outcomes (57). However, the function of CCNB1 remains little known in LUAD. Our studies demonstrated that CCNB1 plays important role in LUAD, which provides novel insight into pathological mechanism involved in LUAD. CDC20, which is an important regulatory protein in the cell cycle, has been identified to be negatively regulated by p53, and used as a potential cancer therapeutic target (58). Several studies showed that the expression of CDC20 was related to the survival in some cancers such as colorectal cancer and hepatocellular carcinoma, and its overexpression predicted a poor prognosis (54,59). TTK is a protein kinase, and its expression is closely associated with cell proliferation (60). Silencing TTK expression inhibits proliferation and progression (61), and the overexpression of TTK promoted breast cancer cell proliferation and conferred a poorer prognosis (62), which indicates that TTK is a risky gene in cancer. RRM2 encodes one of two nonidentical subunits for ribonucleotide reductase that catalyzes the formation of deoxyribonucleotides from ribonucleotides. Several studies have reported that the gene expression was associated with cancers, and it has been identified as potential prognostic marker in patients with NSCLC or breast cancer (63-65). ASPM is the human ortholog of the Drosophila melanogaster abnormal spindle gene (asp), which plays essential role in normal mitotic spindle function in embryonic neuroblasts (66). Recently, some studies showed that ASPM expression was associated with the survival of cancer patients, and its upregulation resulted in a poor prognosis in breast cancer and pancreatic ductal adenocarcinoma (62,67). KIF4A and KIF20A are two members of the kinesin family that are mainly responsible for movement along the microtubules in the cell, which has been demonstrated to be associated with many diseases including cancers (68-71). The other three genes including MELK, TOP2A and TYMS are enzyme-encoding genes, which separately play roles in BPs related to cell cycle regulation, chromosome status, and DNA replication and repair. So far, some published studies have reported that these genes played roles in the initiation and progression of cancers and have been identified as predictors of the survival in cancer patients.
In the 2-gene signature, ADRB2 is a protein coding gene encoding the beta2 adrenergic receptor that belongs to a member of the G protein-coupled receptor (GPCR) superfamily. Many studies have confirmed that several diseases such as asthma (72), obesity (73) and type 2 diabetes (74) were associated with ADRB2. Recently, researchers have found that ADRB2 was significantly correlated with various aspects related to cancer such as prostate cancer (75) and breast cancer (74), which indicates that ADRB2 is related to cell proliferation and apoptosis, tumor growth and metastasis, and angiogenesis. A few studies have so far discovered that ADRB2 activation promoted the proliferation of LC cells (76), and it was identified as a potential independent factor for early-stage NSCLC patients (77). Our results confirmed that the dysregulation of ADRB2 played a key role in LUAD, and high mRNA expression of ADRB2 resulted in a higher OS rate in LUAD patients. Similar to ADRB2, VIPR1 encodes a small neuropeptide that belongs to GPCR. VIPR1 is widely expressed among various tissues and plays key roles in many physiological functions including immune regulation, glycogen metabolism, etc. (78). Previous studies have found that VIPR1 was differentially expressed between cancer and normal tissues in many malignancies (78). For example, VIPR1 was highly expressed in prostate cancer (79), breast cancer (80) and colon cancer (81), which indicates that it functions in malignant tumors. Some studies have also reported that VIPR1 was significantly downregulated in LC (82) and may serve as a molecular target for the diagnosis, prevention and treatment of LC (80). This study further demonstrated that VIPR1 played roles in LUAD, and high mRNA expression of VIPR1 resulted in a higher OS rate in LUAD patients. Interestingly, both ADRB2 and VIPR1 identified in this study are GPCRs that belong to the largest family of cell surface receptors mediating many physiological processes (83). Large numbers of studies have proven that GPCRs play roles in many diseases processes such as human genetic and endocrine diseases (84,85), and GPCRs are often designated as drug targets (86). Despite their broad physiological and pathological functions, as well as acting as favorable sites for drug development, the roles of GPCRs have been underappreciated for a long time in tumor biology (83). With the discovery of more GPCRs associated with cancers, researchers have recognized the importance of the functions of GPCRs in tumor biology and paid more attention to GPCRs in many aspects such as molecular machinery (87-89) and tumor-related drug development (86,90). In this study, we found that two key GPCR genes played roles in LUAD by a comprehensive analysis, which indicates that GPCRs provide important functions in tumor biology. Two identified genes may serve as potential diagnostic and prognostic candidates and pharmacological drug targets in LUAD patients.
In addition, several small molecules were identified as potential therapeutic drugs to combat LUAD in this study. Especially, irinotecan (Drugbank accession number: DB00762) was found in three small-molecule prediction databases. Irinotecan is a derivative of camptothecin that inhibits the action of topoisomerase I and can prevent the religation of the DNA strand by binding to the topoisomerase I-DNA complex and causing double-strand DNA breakage and cell death. Irinotecan, as an antineoplastic enzyme inhibitor, is primarily used in the treatment of colorectal cancer. Recently, irinotecan was also approved for treating advanced pancreatic cancer. In LC, the therapeutic efficacy of irinotecan was evaluated by a large number of trials (91-93). In particular, irinotecan has been proven to be effective as a chemotherapeutic drug against small cell lung cancer (SCLC), and SCLC patients receiving maintenance chemotherapy with irinotecan had a longer survival (94). Currently, a few studies have also explored the treatment effect of irinotecan against NSCLC (95).Our results showed that irinotecan might serve as potential therapeutic drug against LUAD. In addition, other small molecules including vincristine, teniposide, etoposide, methotrexate, podophyllotoxin and others were also predicted in this study. Some of these small molecules such as vincristine and teniposide have so far been used for anti-tumor therapy (96,97), and these small molecules may play certain roles in combating LUAD.
Despite the findings in terms of clinical implications, some limitations should be noted. First, this study is a retrospective study based on a bioinformatics strategy, so the robustness of the prediction value of the gene signature should be further validated by prospective clinical trials. Second, the functional roles of the gene signature should be further elucidated in LUAD.
Conclusions
Taken together, the present study systematically analyzed gene expression data related to LUAD using comprehensive bioinformatics methods and identified some key genes associated with the diagnosis and prognosis of LUAD patients. In addition, some small molecules were predicted to combat LUAD. These findings provide novel insights into the pathological mechanism involved in LUAD and may serve as potential diagnostic and prognostic markers and therapy targets against LUAD.
Acknowledgments
We would like to thank American Journal Experts for their helps in polishing our paper.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist and the MDAR checklist. Available at: https://dx.doi.org/10.21037/tcr-21-526
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-526
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-526). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of The First People’s Hospital of Yunnan Province (No. 2017YY227). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017;7:170070. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Travis WD. Pathology of lung cancer. Clin Chest Med 2002;23:65-81. viii. [Crossref] [PubMed]
- Zagryazhskaya A, Gyuraszova K, Zhivotovsky B. Cell death in cancer therapy of lung adenocarcinoma. Int J Dev Biol 2015;59:119-29. [Crossref] [PubMed]
- Wu K, House L, Liu W, et al. Personalized targeted therapy for lung cancer. Int J Mol Sci 2012;13:11471-96. [Crossref] [PubMed]
- Nesbitt JC, Putnam JB Jr, Walsh GL, et al. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg 1995;60:466-72. [Crossref] [PubMed]
- Walters S, Maringe C, Coleman MP, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax 2013;68:551-64. [Crossref] [PubMed]
- Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816-24. [Crossref] [PubMed]
- Ohba T, Toyokawa G, Osoegawa A, et al. Mutations of the EGFR, K-ras, EML4-ALK, and BRAF genes in resected pathological stage I lung adenocarcinoma. Surg Today 2016;46:1091-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Muller IB, de Langen AJ, Giovannetti E, et al. Anaplastic lymphoma kinase inhibition in metastatic non-small cell lung cancer: clinical impact of alectinib. Onco Targets Ther 2017;10:4535-41. [Crossref] [PubMed]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010;10:760-74. [Crossref] [PubMed]
- Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med 2012;18:349-51. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010;466:869-73. [Crossref] [PubMed]
- Songyang Y, Zhu W, Liu C, et al. Large-scale gene expression analysis reveals robust gene signatures for prognosis prediction in lung adenocarcinoma. PeerJ 2019;7:e6980. [Crossref] [PubMed]
- Saji H, Tsuboi M, Shimada Y, et al. Gene expression profiling and molecular pathway analysis for the identification of early-stage lung adenocarcinoma patients at risk for early recurrence. Oncol Rep 2013;29:1902-6. [Crossref] [PubMed]
- Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One 2008;3:e1651. [Crossref] [PubMed]
- Lu TP, Tsai MH, Lee JM, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev 2010;19:2590-7. [Crossref] [PubMed]
- Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990;323:561-5. [Crossref] [PubMed]
- Liu HY, Zhao H, Li WX. Integrated Analysis of Transcriptome and Prognosis Data Identifies FGF22 as a Prognostic Marker of Lung Adenocarcinoma. Technol Cancer Res Treat 2019;18:1533033819827317. [Crossref] [PubMed]
- Wang L, Meng Y, Zhang QY. LAPTM4B is a novel diagnostic and prognostic marker for lung adenocarcinoma and associated with mutant EGFR. BMC Cancer 2019;19:293. [Crossref] [PubMed]
- Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 2007;8:140. [Crossref] [PubMed]
- Sivakumar S, Lucas FAS, McDowell TL, et al. Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma. Cancer Res 2017;77:6119-30. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Koschützki D, Schreiber F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul Syst Bio 2008;2:193-201. [Crossref] [PubMed]
- Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72. [Crossref] [PubMed]
- Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med 2018;16:264. [Crossref] [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Wang Z, Lachmann A, Keenan AB, et al. L1000FWD: fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics 2018;34:2150-2. [Crossref] [PubMed]
- Cotto KC, Wagner AH, Feng YY, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res 2018;46:D1068-73. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science 1994;266:1821-8. [Crossref] [PubMed]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316-23. [Crossref] [PubMed]
- Giordano TJ, Kuick R, Else T, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res 2009;15:668-76. [Crossref] [PubMed]
- Huang R, Gao L. Identification of potential diagnostic and prognostic biomarkers in non-small cell lung cancer based on microarray data. Oncol Lett 2018;15:6436-42. [Crossref] [PubMed]
- Peng F, Li Q, Niu SQ, et al. ZWINT is the next potential target for lung cancer therapy. J Cancer Res Clin Oncol 2019;145:661-73. [Crossref] [PubMed]
- Woo Seo D, Yeop You S, Chung WJ, et al. Zwint-1 is required for spindle assembly checkpoint function and kinetochore-microtubule attachment during oocyte meiosis. Sci Rep 2015;5:15431. [Crossref] [PubMed]
- Ying H, Xu Z, Chen M, et al. Overexpression of Zwint predicts poor prognosis and promotes the proliferation of hepatocellular carcinoma by regulating cell-cycle-related proteins. Onco Targets Ther 2018;11:689-702. [Crossref] [PubMed]
- Zhou G, Shen M, Zhang Z. ZW10 Binding Factor (ZWINT), a Direct Target of Mir-204, Predicts Poor Survival and Promotes Proliferation in Breast Cancer. Med Sci Monit 2020;26:e921659. [Crossref] [PubMed]
- Wang HJ, Wang L, Lv J, et al. Decreased expression of Zwint-1 is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol 2017;10:10406-12. [PubMed]
- Yang XY, Wu B, Ma SL, et al. Decreased Expression of ZWINT is Associated With Poor Prognosis in Patients With HCC After Surgery. Technol Cancer Res Treat 2018;17:1533033818794190. [Crossref] [PubMed]
- Qian D, Zheng W, Chen C, et al. Roles of CCNB2 and NKX3-1 in Nasopharyngeal Carcinoma. Cancer Biother Radiopharm 2020;35:208-13. [Crossref] [PubMed]
- Qian X, Song X, He Y, et al. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother 2015;74:222-7. [Crossref] [PubMed]
- Bie L, Zhao G, Ju Y, et al. Integrative genomic analysis identifies CCNB1 and CDC2 as candidate genes associated with meningioma recurrence. Cancer Genet 2011;204:536-40. [Crossref] [PubMed]
- Zhuang L, Yang Z, Meng Z. Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in Tumor Tissues Predicted Worse Overall Survival and Disease-Free Survival in Hepatocellular Carcinoma Patients. Biomed Res Int 2018;2018:7897346. [Crossref] [PubMed]
- Yang WX, Pan YY, You CG. CDK1, CCNB1, CDC20, BUB1, MAD2L1, MCM3, BUB1B, MCM2, and RFC4 May Be Potential Therapeutic Targets for Hepatocellular Carcinoma Using Integrated Bioinformatic Analysis. Biomed Res Int 2019;2019:1245072. [Crossref] [PubMed]
- Wang S, Sun H, Zhan X, et al. MicroRNA-718 serves a tumor-suppressive role in non-small cell lung cancer by directly targeting CCNB1. Int J Mol Med 2020;45:33-44. [PubMed]
- Liu D, Xu W, Ding X, et al. Polymorphisms of CCNB1 Associated With the Clinical Outcomes of Platinum-Based Chemotherapy in Chinese NSCLC Patients. J Cancer 2017;8:3785-94. [Crossref] [PubMed]
- Kidokoro T, Tanikawa C, Furukawa Y, et al. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008;27:1562-71. [Crossref] [PubMed]
- Wu WJ, Hu KS, Wang DS, et al. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med 2013;11:142. [Crossref] [PubMed]
- Mills GB, Schmandt R, McGill M, et al. Expression of TTK, a novel human protein kinase, is associated with cell proliferation. J Biol Chem 1992;267:16000-6. [Crossref] [PubMed]
- Chen S, Wang J, Wang L, et al. Silencing TTK expression inhibits the proliferation and progression of prostate cancer. Exp Cell Res 2019;385:111669. [Crossref] [PubMed]
- Tang J, Lu M, Cui Q, et al. Overexpression of ASPM, CDC20, and TTK Confer a Poorer Prognosis in Breast Cancer Identified by Gene Co-expression Network Analysis. Front Oncol 2019;9:310. [Crossref] [PubMed]
- Putluri N, Maity S, Kommagani R, et al. Pathway-centric integrative analysis identifies RRM2 as a prognostic marker in breast cancer associated with poor survival and tamoxifen resistance. Neoplasia 2014;16:390-402. [Crossref] [PubMed]
- Wang L, Meng L, Wang XW, et al. Expression of RRM1 and RRM2 as a novel prognostic marker in advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol 2014;35:1899-906. [Crossref] [PubMed]
- Zhao H, Zhang H, Du Y, et al. Prognostic significance of BRCA1, ERCC1, RRM1, and RRM2 in patients with advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol 2014;35:12679-88. [Crossref] [PubMed]
- Bond J, Roberts E, Mochida GH, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet 2002;32:316-20. [Crossref] [PubMed]
- Tian X, Wang N. Upregulation of ASPM, BUB1B and SPDL1 in tumor tissues predicts poor survival in patients with pancreatic ductal adenocarcinoma. Oncol Lett 2020;19:3307-15. [Crossref] [PubMed]
- Xie F, He C, Gao S, et al. KIF20A silence inhibits the migration, invasion and proliferation of non-small cell lung cancer and regulates the JNK pathway. Clin Exp Pharmacol Physiol 2020;47:135-42. [Crossref] [PubMed]
- Xiong M, Zhuang K, Luo Y, et al. KIF20A promotes cellular malignant behavior and enhances resistance to chemotherapy in colorectal cancer through regulation of the JAK/STAT3 signaling pathway. Aging (Albany NY) 2019;11:11905-21. [Crossref] [PubMed]
- Han Q, Han C, Liao X, et al. Prognostic value of Kinesin-4 family genes mRNA expression in early-stage pancreatic ductal adenocarcinoma patients after pancreaticoduodenectomy. Cancer Med 2019;8:6487-502. [Crossref] [PubMed]
- Hou G, Dong C, Dong Z, et al. Upregulate KIF4A Enhances Proliferation, Invasion of Hepatocellular Carcinoma and Indicates poor prognosis Across Human Cancer Types. Sci Rep 2017;7:4148. [Crossref] [PubMed]
- Toraih EA, Hussein MH, Ibrahim A, et al. Beta2-adrenergic receptor variants in children and adolescents with bronchial asthma. Front Biosci (Elite Ed) 2019;11:61-78. [PubMed]
- Mitra SR, Tan PY, Amini F. Association of ADRB2 rs1042713 with Obesity and Obesity-Related Phenotypes and Its Interaction with Dietary Fat in Modulating Glycaemic Indices in Malaysian Adults. J Nutr Metab 2019;2019:8718795. [Crossref] [PubMed]
- Connor A, Baumgartner RN, Kerber RA, et al. ADRB2 G-G haplotype associated with breast cancer risk among Hispanic and non-Hispanic white women: interaction with type 2 diabetes and obesity. Cancer Causes Control 2012;23:1653-63. [Crossref] [PubMed]
- Kulik G. ADRB2-Targeting Therapies for Prostate Cancer. Cancers (Basel) 2019;11:358. [Crossref] [PubMed]
- Hu P, He J, Liu S, et al. β2-adrenergic receptor activation promotes the proliferation of A549 lung cancer cells via the ERK1/2/CREB pathway. Oncol Rep 2016;36:1757-63. [Crossref] [PubMed]
- Yazawa T, Kaira K, Shimizu K, et al. Prognostic significance of β2-adrenergic receptor expression in non-small cell lung cancer. Am J Transl Res 2016;8:5059-70. [PubMed]
- Lu S, Lu H, Jin R, et al. Promoter methylation and H3K27 deacetylation regulate the transcription of VIPR1 in hepatocellular carcinoma. Biochem Biophys Res Commun 2019;509:301-5. [Crossref] [PubMed]
- Xie Y, Wolff DW, Lin MF, et al. Vasoactive intestinal peptide transactivates the androgen receptor through a protein kinase A-dependent extracellular signal-regulated kinase pathway in prostate cancer LNCaP cells. Mol Pharmacol 2007;72:73-85. [Crossref] [PubMed]
- Moody TW, Gozes I. Vasoactive intestinal peptide receptors: a molecular target in breast and lung cancer. Curr Pharm Des 2007;13:1099-104. [Crossref] [PubMed]
- Liu S, Zeng Y, Li Y, et al. VPAC1 overexpression is associated with poor differentiation in colon cancer. Tumour Biol 2014;35:6397-404. [Crossref] [PubMed]
- Mlakar V, Strazisar M, Sok M, et al. Oligonucleotide DNA microarray profiling of lung adenocarcinoma revealed significant downregulation and deletions of vasoactive intestinal peptide receptor 1. Cancer Invest 2010;28:487-94. [Crossref] [PubMed]
- Bar-Shavit R, Maoz M, Kancharla A, et al. G Protein-Coupled Receptors in Cancer. Int J Mol Sci 2016;17:1320. [Crossref] [PubMed]
- Lania AG, Mantovani G, Spada A. Mechanisms of disease: Mutations of G proteins and G-protein-coupled receptors in endocrine diseases. Nat Clin Pract Endocrinol Metab 2006;2:681-93. [Crossref] [PubMed]
- Thompson MD, Percy ME, McIntyre Burnham W, et al. G protein-coupled receptors disrupted in human genetic disease. Methods Mol Biol 2008;448:109-37. [Crossref] [PubMed]
- Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov 2011;10:47-60. [Crossref] [PubMed]
- Murugan AK, Qasem E, Al-Hindi H, et al. GPCR-mediated PI3K pathway mutations in pediatric and adult thyroid cancer. Oncotarget 2019;10:4107-24. [Crossref] [PubMed]
- Lappano R, Jacquot Y, Maggiolini M. GPCR Modulation in Breast Cancer. Int J Mol Sci 2018;19:3840. [Crossref] [PubMed]
- Nogués L, Palacios-García J, Reglero C, et al. G protein-coupled receptor kinases (GRKs) in tumorigenesis and cancer progression: GPCR regulators and signaling hubs. Semin Cancer Biol 2018;48:78-90. [Crossref] [PubMed]
- Liu Y, An S, Ward R, et al. G protein-coupled receptors as promising cancer targets. Cancer Lett 2016;376:226-39. [Crossref] [PubMed]
- Arnold SM, Chansky K, Baggstrom MQ, et al. Phase II Trial of Carfilzomib Plus Irinotecan in Patients With Small-cell Lung Cancer Who Have Progressed on Prior Platinum-based Chemotherapy. Clin Lung Cancer 2020;21:357-364.e7. [Crossref] [PubMed]
- Kondo R, Watanabe S, Shoji S, et al. A Phase II Study of Irinotecan for Patients with Previously Treated Small-Cell Lung Cancer. Oncology 2018;94:223-32. [Crossref] [PubMed]
- Misumi Y, Okamoto H, Sasaki J, et al. Phase I/II study of induction chemotherapy using carboplatin plus irinotecan and sequential thoracic radiotherapy (TRT) for elderly patients with limited-disease small-cell lung cancer (LD-SCLC): TORG 0604. BMC Cancer 2017;17:377. [Crossref] [PubMed]
- Yagi Y, Kim YH, Tajima N, et al. Long survival of a small-cell lung cancer patient who received maintenance chemotherapy with irinotecan. Case Rep Oncol 2013;6:569-73. [Crossref] [PubMed]
- Han N, Liu ZW, Wang J, et al. Clinical study of irinotecan plus cisplatin for advanced non-small cell lung cancer. Nan Fang Yi Ke Da Xue Xue Bao 2010;30:349-50. [PubMed]
- Lin FZ, Wang SC, Hsi YT, et al. Celastrol induces vincristine multidrug resistance oral cancer cell apoptosis by targeting JNK1/2 signaling pathway. Phytomedicine 2019;54:1-8. [Crossref] [PubMed]
- Yan J, Sun J, Zeng Z. Teniposide ameliorates bone cancer nociception in rats via the P2X7 receptor. Inflammopharmacology 2018;26:395-402. [Crossref] [PubMed]


