Characterization of genomic clones by targeted deep sequencing of ctDNA to monitor liver cancer
Introduction
Circulating tumor DNA (ctDNA) is a type of cell-free extracellular DNA that is found in body fluids, such as blood (1). ctDNA exists in the form of DNA-protein complexes or free DNA (1). Some studies have shown that the origin of ctDNA is related to tumor cells [tumor cells or circulating tumor cells (CTCs)]. ctDNA is formed when these cells are released into the circulatory system due to shedding or apoptosis. Circulating nucleic acids were discovered by Mandel and Metais as early as 1947 (2), and Shapiro et al. showed that peripheral blood serum levels in tumor patients are significantly higher than those in normal subjects (3). Researchers have also detected oncogene mutations in the plasma and serum of tumor patients, indicating that the mutation spectrum of ctDNA is consistent with the mutation spectrum of the primary tumor. Due to the potential of ctDNA in tumor prognosis and recurrence monitoring as well as its noninvasive advantages, it has become a hot research topic in recent years (4).
Current methods for detecting tumorigenesis, recurrence and metastasis include tumor imaging methods, such as CT and PET-CT, and serological markers, such as alpha-fetoprotein (AFP). However, due to their low specificity and sensitivity, traditional serological markers (such as AFP) cannot meet clinical needs. ctDNA, which is present in peripheral blood, has great potential in the diagnosis of tumors and the monitoring of therapeutic effects. ctDNA is easy to amplify and can be detected by highly sensitive instruments. The “liquid biopsy” of ctDNA allows an instant understanding of what happens to tumor patients. Compared to traditional tissue biopsy, ctDNA detection can be repeated and resampled, which is more suitable for patients who need full-course efficacy evaluation of disease progression. ctDNA can also be used as an important monitoring marker for the evaluation of therapeutic effects and clinical follow-up after treatment.
Next-generation sequencing (NGS) provides a simultaneous, massive parallel and high-throughput method to detect thousands of mutations in cancer patients. With the development of personalized treatment, comprehensive characterization of genetic alterations using next-generation sequencing has been demonstrated to be helpful to guide the selection of therapy and predict prognosis. Due to the low concentration of ctDNA, many researchers have performed high-throughput sequencing methods to study ctDNA. Using massive parallel sequencing, it has been reported that ctDNA is an effective marker and is associated with many types of human cancers. For example, Leary et al. proposed a highly sensitive and widely used method for improving the clinical management of cancer patients with personalized biomarkers (5). Maniesh van der Vaart et al. (6) used the GSFLX sequencing platform (454) to analyze ctDNA extracted from 12 prostate cancer patients. Chan et al. (7) used a shotgun method to detect ctDNA in four hepatocellular carcinoma patients and one patient with bilateral breast and ovarian cancer in a noninvasive manner, and they found copy number variations associated with cancer. These studies clarify the feasibility of this method in studying tumor characteristics and its utility in detecting, monitoring, and studying cancer. In a study of 30 women with metastatic breast cancer who underwent therapy, Dawson et al. (8) compared the performance of ctDNA, CA15-3 and CTCs. Compared to tumor imaging findings, the results indicated that ctDNA is a biomarker with high specificity and high sensitivity. Research on ctDNA has developed rapidly and has become a research hotspot in the field of cancer (4), indicating its high potential in tumor monitoring and improvement of prognosis.
Liver cancer is one of the most common causes of cancer-associated mortalities in China (8), and more than 350,000 patients die of liver cancer every year (9). Thus, it is necessary to develop a noninvasive detection method for liver cancer. There are few approved targeted drugs for the treatment of liver cancer. Most patients with liver cancer (especially those with metastatic lesions) need to be treated with chemotherapy. Studies have shown that targeted therapy is safer and more effective in the treatment of cancer patients (1,10,11). Some studies have shown that ctDNA can be used (8,12) to detect specific variants in tumor patients quantified in a noninvasive manner. Cancer patients can also be monitored in real time through the detection of ctDNA, which may help to detect the progress of the disease, adjust the treatment plan, and achieve the purpose of prolonging the survival of the patients. However, in liver cancer patients, ctDNA has not been fully analyzed or investigated. To detect clinically relevant mutations in liver cancer patients, we present the first study using targeted NGS technology on a BGISEQ-500 sequencer.
In this study, we developed a new panel containing 378 liver cancer-related genes. Using the BGISEQ-500 platform, we first tested the performance of this panel in detecting clinically relevant mutations. Using this panel, we then detected 266 clinically relevant mutations in the pretreatment plasma samples of 34 clinical cases. We also investigated the clonal changes before and after surgery in 29 liver cancer patients. The results showed that ctDNA can be used in the monitoring of liver cancer patients. This study, to the best of our knowledge, is the first to detect clinically relevant alterations in liver cancer in the Chinese population using the BGISEQ-500 platform. We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/tcr-21-1005).
Methods
Virtual plasma samples and patients with liver cancer
To calculate the accuracy and stability of this technology, virtual plasma samples harboring known mutations of gradient allelic frequency were obtained. First, plasma samples of 10 normal controls were collected and mixed to prepare plasma basal fluid (DY). Then, 15 tumor DNA samples with known mutations (7 from Horizon and 8 from Cobioer) were fragmented to 160 bp (Table S1). After dilution, the DNA fragment was then mixed with DY at different mutation frequencies to form a virtual plasma sample (ZJ). ZJ contains 15 different types of mutations, including SNV, indel, and gene fusion. As shown in Table S1, the final frequency span of the mutation in ZJ was 0.23–10%.
The present study consisted of 34 liver cancer patients from The Central Hospital of Wuhan. Samples of liver cancer patients were collected and analyzed between September 2017 and January 2018. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of BGI (BGI-IRB 15136). Informed consent was obtained from all participants.
DNA extraction and quantification
Genomic DNA was isolated from virtual plasma samples (ZJ) harboring known mutations and blood samples before and after operation or biopsy. Following standard protocols, the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) was used to extract total genomic DNA from ZJ and peripheral blood lymphocytes (PBLs) of the patients. Plasma DNA of liver cancer patients was extracted by a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). DNA quality and quantity were assessed using a Nanodrop and Qubit (Thermo Fisher Scientific).
Library preparation, targeted capture, and sequencing
In this study, the DNA samples were sequenced by the BGISEQ-500 platform. Following the standard protocol provided by BGI (BGI-Shenzhen), we performed library preparation, sequence capture, and sequencing (BGISEQ-500RS High-throughput sequencing kit, PE100, V3.0, MGI Tech Co., Ltd.) for all the plasma samples. In brief, 20 ng of DNA was used for end repair and A-tailing. The fragments were then ligated to adapters (23 °C for 1 h), and the ligation products were purified and amplified. After purification, 8 libraries were pooled together and hybridized to the capture array (65 °C for 16 h). Following the manufacturer’s instructions (MGI Tech Co., Ltd.), the hybridized capture array was washed and eluted. The circularization process was performed for each eluted library, and the DNBs were loaded into a sequencing chip for paired-end (110 bp) sequencing.
Bioinformatics analysis
After image analysis and base calling by the BGISEQ-500 platform, FASTQ data were generated. The pipeline of bioinformatics analysis mainly included four steps. First, SOAPnuke was used to filter reads with low quality and adapter sequences. Second, BWA-mem was used to generate bam files. Sorted BAM files were then generated using Picard, which was also used to remove PCR-derived duplications. Third, MuTect2 software was used to detect somatic SNVs and indels. CopyWriteR was used to identify somatic CNVs, and somatic fusions were detected by self-developed software. Fourth, all the above identified mutations were annotated by our own knowledge database (BGI-CKD) for further analysis.
Statistical analysis
No specific statistical analysis was performed in the current study.
Results
Study design and patient enrollment
In the present, we developed a targeted next-generation sequencing method using the BGISEQ-500 platform to detect clinically relevant mutations in clinical cases of liver cancer. We first collected informed consent from all participants. A total of 34 patients were recruited from The Central Hospital of Wuhan in this study (Table 1). The AFP levels were measured at different time points. Moreover, a virtual tumor sample harboring known mutations of gradient allelic frequency (0.23–10%) was analyzed three times (Table S1). The study and the protocols used were approved by the Institutional Ethics Committee of BGI-Shenzhen (BGI-IRB 15136).
Table 1
| Number | Sample name | Time point | Concentration (ng/uL) | Total (ng) | Sequencing depth | Coverage (%) |
|---|---|---|---|---|---|---|
| 1 | LZ01-L1 | Before surgery | 24.4 | 1,220 | 567.95 | 99.86 |
| LZ01-P1 | 1.5 | 67.5 | 2,416.04 | 99.99 | ||
| LZ01-L2 | One week after surgery | 22.4 | 1,120 | 656.33 | 99.88 | |
| LZ01-P2 | 1.28 | 57.6 | 1,634.06 | 99.96 | ||
| LZ01-L3 | One month after surgery | 27.2 | 1,360 | 717.75 | 99.9 | |
| LZ01-P3 | 0.742 | 33.39 | 1,537.67 | 99.94 | ||
| 2 | LZ02-L1 | Before surgery | 39.6 | 1,980 | 642.63 | 99.88 |
| LZ02-P1 | 0.524 | 23.58 | 776.69 | 99.9 | ||
| LZ02-L2 | One week after surgery | 48.4 | 2,420 | 682.65 | 99.92 | |
| LZ02-P2 | 0.844 | 37.98 | 2,099.44 | 99.98 | ||
| 3 | LZ03-L1 | Before surgery | 60 | 3,000 | 708.17 | 99.87 |
| LZ03-P1 | 1.77 | 79.65 | 1,553.43 | 99.92 | ||
| LZ03-L2 | One week after surgery | 34 | 1,700 | 678.36 | 99.88 | |
| LZ03-P2 | 2.64 | 118.8 | 2841 | 99.95 | ||
| LZ03-L3 | One month after surgery | 26.4 | 1,320 | 690.83 | 99.88 | |
| LZ03-P3 | 0.892 | 40.14 | 1,482.67 | 99.93 | ||
| 4 | LZ04-L1 | Before surgery | 13.7 | 685 | 628.8 | 99.84 |
| LZ04-P1 | 0.596 | 26.82 | 1,613.45 | 99.99 | ||
| LZ04-L2 | One week after surgery | 33.4 | 1670 | 370.76 | 99.83 | |
| LZ04-P2 | 3.34 | 150.3 | 1,765.08 | 99.95 | ||
| LZ04-L3 | One month after surgery | 14.4 | 720 | 584.6 | 99.87 | |
| LZ04-P3 | 0.6 | 27 | 1,555.47 | 99.96 | ||
| 5 | LZ05-L1 | Before surgery | 53 | 2650 | 488.69 | 99.87 |
| LZ05-P1 | 0.668 | 30.06 | 1,938.95 | 99.98 | ||
| LZ05-L2 | One week after surgery | 73.4 | 3670 | 676.3 | 99.88 | |
| LZ05-P2 | 4.4 | 198 | 2,556.93 | 99.98 | ||
| 6 | LZ06-L1 | Before surgery | 11.1 | 555 | 545.51 | 99.82 |
| LZ06-P1 | 1.22 | 54.9 | 1,164.22 | 99.94 | ||
| LZ06-L2 | One week after surgery | 27.2 | 1,360 | 504.67 | 99.81 | |
| LZ06-P2 | 1.34 | 60.3 | 1,465.36 | 99.94 | ||
| 7 | LZ07-L1 | Before surgery | 59.6 | 2,980 | 534.51 | 99.8 |
| LZ07-P1 | 1.9 | 85.5 | 1,413.4 | 99.95 | ||
| LZ07-L2 | One week after surgery | 94.8 | 4740 | 534.32 | 99.81 | |
| LZ07-P2 | 1.82 | 81.9 | 1,802.31 | 99.93 | ||
| 8 | LZ08-L1 | 0.3-month after surgery | 28 | 1,400 | 613.87 | 99.87 |
| LZ08-P1 | 0.506 | 22.77 | 659.42 | 99.86 | ||
| LZ08-L2 | 0.57-month after surgery | 31.6 | 1,580 | 584.39 | 99.84 | |
| LZ08-P2 | 7.3 | 328.5 | 3,987.2 | 99.95 | ||
| LZ08-L3 | 0.7-month after surgery | 49 | 2450 | 553.75 | 99.87 | |
| LZ08-P3 | 2.78 | 125.1 | 1,889.31 | 99.92 | ||
| 9 | LZ09-L1 | Before surgery | 110 | 5,500 | 645.79 | 99.9 |
| LZ09-P1 | 0.584 | 26.28 | 1,209.81 | 99.98 | ||
| LZ09-L2 | One week after surgery | 62.6 | 3,130 | 596.6 | 99.89 | |
| LZ09-P2 | 1.93 | 86.85 | 1,584.38 | 100 | ||
| LZ09-L3 | One month after surgery | 65 | 2,600 | 822.08 | 99.89 | |
| LZ09-P3 | 0.308 | 15.4 | 1,630.99 | 99.96 | ||
| 10 | LZ10-L1 | Before surgery | 115 | 5,750 | 628.14 | 99.93 |
| LZ10-P1 | 1.18 | 53.1 | 1,820.79 | 99.99 | ||
| LZ10-L2 | One week after surgery | 49 | 2,450 | 575.16 | 99.89 | |
| LZ10-P2 | 17.3 | 778.5 | 3,375.8 | 99.99 | ||
| 11 | LZ11-L1 | 0.2-month after surgery | 52.6 | 2,630 | 637.02 | 99.86 |
| LZ11-P1 | 0.822 | 36.99 | 1,755.98 | 99.95 | ||
| LZ11-L2 | 0.5-month after surgery | 67.4 | 3,370 | 608.57 | 99.94 | |
| LZ11-P2 | 1.45 | 65.25 | 1,865.68 | 99.96 | ||
| 12 | LZ12-L1 | Before surgery | 62.4 | 3,120 | 611.33 | 99.87 |
| LZ12-P1 | 0.644 | 28.98 | 1,118.99 | 99.9 | ||
| LZ12-L2 | One week after surgery | 23.4 | 1,170 | 615.92 | 99.89 | |
| LZ12-P2 | 0.724 | 32.58 | 1,623.62 | 99.94 | ||
| LZ12-L3 | One month after surgery | 16.2 | 648 | 685.2 | 99.89 | |
| LZ12-P3 | 0.34 | 17 | 1,575.76 | 99.95 | ||
| 13 | LZ13-L1 | Before surgery | 88.6 | 4,430 | 709.52 | 99.9 |
| LZ13-P1 | 1.71 | 76.95 | 2,793.03 | 99.99 | ||
| LZ13-L2 | One week after surgery | 22.8 | 1,140 | 589.9 | 99.9 | |
| LZ13-P2 | 3.8 | 171 | 1,802.37 | 99.99 | ||
| 14 | LZ14-L1 | Before surgery | 33.6 | 1,680 | 656.36 | 99.89 |
| LZ14-P1 | 0.68 | 30.6 | 1,171.39 | 99.96 | ||
| LZ14-L2 | One week after surgery | 58.4 | 2,920 | 601.72 | 99.88 | |
| LZ14-P2 | 1.31 | 58.95 | 1,614.49 | 99.95 | ||
| LZ14-L3 | One month after surgery | 24.2 | 1,210 | 550.76 | 99.88 | |
| LZ14-P3 | 1.39 | 62.55 | 1,694.25 | 100 | ||
| 15 | LZ15-L1 | 4.13 months after surgery | 15 | 750 | 421.73 | 99.88 |
| LZ15-P1 | 0.672 | 30.24 | 1,334.97 | 99.99 | ||
| LZ15-L2 | 4.33 months after surgery | 100 | 5,000 | 350.85 | 99.88 | |
| LZ15-P2 | 3.3 | 148.5 | 2,435.09 | 99.99 | ||
| 16 | LZ16-L1 | Before surgery | 91.8 | 4,590 | 481.87 | 99.92 |
| LZ16-P1 | 0.556 | 25.02 | 539.92 | 99.95 | ||
| LZ16-L2 | One week after surgery | 21.8 | 1,090 | 537.63 | 99.88 | |
| LZ16-P2 | 0.948 | 42.66 | 1,191.35 | 99.96 | ||
| 17 | LZ17-L1 | Before surgery | 45.6 | 2,280 | 497.08 | 99.88 |
| LZ17-P1 | 0.764 | 34.38 | 1,048.74 | 100 | ||
| LZ17-L2 | One week after surgery | 35 | 1,750 | 561.99 | 99.89 | |
| LZ17-P2 | 1.65 | 74.25 | 1994.74 | 100 | ||
| LZ17-L3 | One month after surgery | NA | NA | 875.47 | 99.89 | |
| LZ17-P3 | 0.48 | 24 | 827.99 | 99.89 | ||
| 18 | LZ18-L1 | Before surgery | 17 | 850 | 481.18 | 99.89 |
| LZ18-P1 | 0.998 | 44.91 | 1,648.15 | 100 | ||
| LZ18-L2 | One week after surgery | 112 | 5,600 | 490.28 | 99.88 | |
| LZ18-P2 | 1.27 | 57.15 | 2,141.24 | 100 | ||
| 19 | LZ19-L1 | Before surgery | 19 | 950 | 480.49 | 99.89 |
| LZ19-P1 | 0.698 | 31.41 | 906.08 | 99.98 | ||
| LZ19-L2 | One week after surgery | 88.6 | 4,430 | 508.83 | 99.88 | |
| LZ19-P2 | 2.48 | 111.6 | 2,182.71 | 100 | ||
| LZ19-L3 | One month after surgery | 20.2 | 808 | 632.48 | 99.84 | |
| LZ19-P3 | 0.662 | 33.1 | 1,699.08 | 99.95 | ||
| 20 | LZ20-L1 | Before surgery | 32.6 | 1,630 | 596.67 | 99.92 |
| LZ20-P1 | 0.764 | 34.38 | 1,634.25 | 99.99 | ||
| LZ20-L2 | One week after surgery | 110 | 5,500 | 720.95 | 99.88 | |
| LZ20-P2 | 1.47 | 66.15 | 2,829.23 | 99.99 | ||
| 21 | LZ21-L1 | Before surgery | 38.4 | 1,920 | 697.29 | 99.88 |
| LZ21-P1 | 0.576 | 25.92 | 1,013.25 | 99.96 | ||
| LZ21-L2 | One week after surgery | 73 | 3,650 | 752.39 | 99.86 | |
| LZ21-P2 | 0.936 | 42.12 | 1,271.58 | 99.99 | ||
| 22 | LZ22-L1 | 5.6 months after surgery | 38.6 | 1,930 | 685.55 | 99.89 |
| LZ22-P1 | 0.856 | 38.52 | 1,506.22 | 99.98 | ||
| LZ22-L2 | 5.67 months after surgery | 45 | 2,250 | 820.42 | 99.92 | |
| LZ22-P2 | 1.57 | 70.65 | 2,643.36 | 100 | ||
| LZ22-L3 | 8.7 months after surgery | 17.7 | 708 | 826 | 99.89 | |
| LZ22-P3 | 0.68 | 34 | 1,839.93 | 99.96 | ||
| 23 | LZ23-L1 | Before surgery | 31.4 | 1,570 | 786.04 | 99.84 |
| LZ23-P1 | 0.548 | 24.66 | 937.14 | 99.86 | ||
| LZ23-L2 | One week after surgery | 123 | 6150 | 750.21 | 99.92 | |
| LZ23-P2 | 1.92 | 86.4 | 1,914.37 | 99.98 | ||
| 24 | LZ24-L1 | Before surgery | NA | NA | 952.3 | 99.9 |
| LZ24-P1 | NA | NA | 475.42 | 99.87 | ||
| LZ24-L2 | One week after surgery | NA | NA | 824.67 | 99.89 | |
| LZ24-P2 | 1.21 | 60.5 | 2,590.66 | 99.99 | ||
| LZ24-L3 | One month after surgery | 44 | 1,760 | 617.35 | 99.87 | |
| LZ24-P3 | 0.548 | 27.4 | 3,356.69 | 99.99 | ||
| 25 | LZ25-L1 | Before surgery | 15.8 | 632 | 690.56 | 99.9 |
| LZ25-P1 | NA | NA | 2,342.37 | 99.96 | ||
| LZ25-L2 | One week after surgery | 24 | 960 | 617.74 | 99.86 | |
| LZ25-P2 | NA | NA | 478.79 | 99.92 | ||
| LZ25-L3 | One month after surgery | NA | NA | 798.17 | 99.87 | |
| LZ25-P3 | 0.872 | 43.6 | 2,509.11 | 99.96 | ||
| 26 | LZ26-L1 | Before surgery | 19.9 | 796 | 698.19 | 99.89 |
| LZ26-P1 | NA | NA | 2,039.71 | 99.94 | ||
| LZ26-L2 | One week after surgery | 28 | 1,120 | 690.76 | 99.89 | |
| LZ26-P2 | NA | NA | 1,670.65 | 99.96 | ||
| 27 | LZ27-L1 | Before surgery | 58.4 | 2,920 | 632.41 | 99.82 |
| LZ27-P1 | 1.03 | 46.35 | 2,350.51 | 99.99 | ||
| LZ27-L3 | One month after surgery | 18.5 | 740 | 709.72 | 99.88 | |
| LZ27-P3 | 0.688 | 34.4 | 1,530.24 | 99.92 | ||
| 28 | LZ28-L1 | Before surgery | 47.2 | 1,888 | 958.64 | 99.9 |
| LZ28-P1 | 0.304 | 15.2 | 136.15 | 99.59 | ||
| LZ28-L2 | One week after surgery | 29.6 | 1,184 | 601.5 | 99.87 | |
| LZ28-P2 | 1.52 | 76 | 2,077.22 | 99.94 | ||
| LZ28-L3 | One month after surgery | 72.4 | 2,896 | 874.11 | 99.9 | |
| LZ28-P3 | 0.924 | 46.2 | 2,082.47 | 99.96 | ||
| 29 | LZ29-L1 | Before surgery | 25.4 | 1,016 | 636.14 | 99.86 |
| LZ29-P1 | 0.152 | 7.6 | 1,332.68 | 99.93 | ||
| LZ29-L2 | One week after surgery | 94.8 | 3,792 | 763.4 | 99.86 | |
| LZ29-P2 | 0.624 | 3,1.2 | 1,806.1 | 99.94 | ||
| 30 | LZ30-L1 | Before surgery | 4.58 | 183.2 | 235.27 | 99.77 |
| LZ30-P1 | 0.626 | 31.3 | 3,220.81 | 99.98 | ||
| LZ30-L2 | 0.3-month after surgery, | 7.58 | 303.2 | 515.32 | 99.84 | |
| LZ30-P2 | 0.52 | 26 | 1,567.56 | 99.93 | ||
| 31 | LZ31-L1 | Before surgery | 70.4 | 2,816 | 813.56 | 99.9 |
| LZ31-P1 | 0.884 | 44.2 | 3,598.54 | 99.96 | ||
| LZ31-L2 | One week after surgery | NA | NA | 908.48 | 99.9 | |
| LZ31-P2 | 0.304 | 15.2 | 1,280.98 | 99.92 | ||
| 32 | LZ32-L1 | Before surgery | 21.4 | 856 | 511.01 | 99.84 |
| LZ32-P1 | 0.204 | 10.2 | 2,245.53 | 99.98 | ||
| LZ32-L2 | One week after surgery | 70 | 2,800 | 729.82 | 99.88 | |
| LZ32-P2 | 0.876 | 43.8 | 3,583.51 | 99.99 | ||
| 33 | LZ33-L1 | Before surgery | 28.8 | 1,152 | 605.47 | 99.89 |
| LZ33-P1 | 4.86 | 243 | 2,098.68 | 99.98 | ||
| LZ33-L2 | One week after surgery | 61.8 | 2,472 | 776.44 | 99.92 | |
| LZ33-P2 | 0.534 | 26.7 | 2,456.37 | 99.95 | ||
| 34 | LZ34-L1 | Before surgery | 43.4 | 1,736 | 671.4 | 99.87 |
| LZ34-P1 | 0.242 | 12.1 | 1,263.81 | 99.96 | ||
| LZ34-L2 | One week after surgery | 28.4 | 1,136 | 739.56 | 99.9 | |
| LZ34-P2 | 0.638 | 31.9 | 2,517.93 | 99.92 |
-L, peripheral blood lymphocytes (PBLs) sample; -P, plasma sample.
To capture the exons of 378 known pathogenic genes (spanning 1,594,704 bp), we designed an array-based chip associated with liver cancer and other common solid tumors. All 378 genes are shown in Table S2. The exons and the 50-bp intron-flanking regions of the 378 genes were included in the designed array. The 378 clinically related genes were identified according to the following criteria: (I) top 20 genes are listed in TCGA, COSMIC and ICGC database; (II) the genes/sites have been previously published to be related to liver cancer in research papers (13-24); (III) genomic alterations in the genes have been demonstrated to be associated with approved cancer therapies in the FDA (www.fda.gov) and NCI-match web pages (https://www.cancer.gov/); and (IV) the NCCN guideline (2017. V1) has clearly indicated that variants in the genes are related to liver cancer.
In the present study, we developed a targeted next-generation sequencing method to detect clinically relevant mutations in 378 liver cancer-related genes using the BGISEQ-500 platform. First, we screened a virtual tumor sample carrying known variants using targeted NGS technology to validate the accuracy and stability of this technology. Second, we detected 266 somatic mutations in the pretreatment plasma samples of the 34 liver cancer patients using this method. Finally, we evaluated ctDNA levels in monitoring tumor burden in liver cancer patients.
Evaluation of sensitivity and stability
Successful extraction of plasma DNA is a prerequisite for subsequent analysis and is important for the accuracy of subsequent analyses. Due to the low concentration of ctDNA in plasma, we first evaluated the accuracy and stability of high-throughput sequencing in detecting ctDNA by constructing virtual tumor samples (ZJs) containing known mutations.
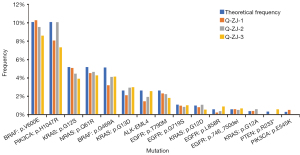
To test the accuracy and stability of our method, virtual plasma samples (ZJs) harboring known mutations (Table S1) of gradient allelic frequency were analyzed (Table S3). Positive cell lines harboring known actionable mutations (obtained from Horizon and Cobioer) were mixed with DNA from 10 healthy controls (DY) and diluted to different allelic frequencies ranging from 0.23% to 10% (ZJ) (Table S1). These virtual tumor samples were used as positive controls and sequenced repeatedly (3 times). As a result, for allelic frequencies of more than 0.5%, 100% (36/36) of the SNV/indel/gene fusions were detected (Figure 1). For allelic frequencies of 0.23–0.31, 44.4% (4/9) of all the known mutations were detected. These results showed that this method detects all variants with an allelic frequency of at least 0.5%, indicating the high sensitivity of this technology.
Targeted region sequencing and data analysis
In this study, 34 patients with liver cancer were sequenced. After removing low-quality reads, BWA mem (Burrows Wheeler Aligner) was used in the alignment process. Read quality control (QC) and alignment QC were performed. We then used Picard tools to remove duplicate reads. Single nucleotide variant (SNV) and indel (short insertions and deletions) detection was performed by Mutect2 software. CNV and gene fusion detection was also performed.
A total of 13,754,633,272 raw reads were generated for all the plasma samples. The sequencing read length was 110 base pairs. The mean sequencing depth for the PBL samples was 642.3-fold after removal of duplications, and the mean sequencing depth for the plasma samples was 1,814.09-fold after removal of duplications (Table 1). For the sequencing coverage of all the plasma samples, an average of 99.35% of the target region was covered by more than 100-fold, and an average of 99.80% of the target region was covered by more than 30-fold. A total of 99.90% of the target region was successfully covered with a depth of more than 10-fold. These results indicated that this method is reliable in detecting clinically relevant mutations.
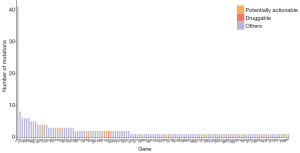
USP6 (15.41% of all mutations), TTN (3.01%), CARD11 (2.26%), SYNE2 (2.26%), CSMD3 (2.26%) and ROS1 (2.26%) were the most frequently mutated tumor genes in the pretreatment plasma samples (Figure 2). Overall, 266 mutations were identified in the pretreatment plasma samples of the 34 liver cancer patients.
Detection of clinically relevant mutations in pretreatment ctDNA
The following criteria were used to identify clinically relevant mutations: (I) the mutations were nonsynonymous variants; (II) the allele frequency of the mutation was more than 1%; (III) the mutations were in key cancer pathways or in the COSMIC database; and (IV) the mutations were reported ≥2 times (25). The potentially actionable mutations were then annotated by our own knowledge database (BGI Cancer Knowledge Database, BGI-CKD). BGI-CKD is a database containing potential clinical implications, FDA-approved drug/therapy targets, or potentially actionable targets that are under active clinical trials.
After annotation, 24 potentially actionable mutations were detected in the pretreatment plasma samples of the liver cancer patients. Only 2 mutations were identified to be targets of approved drugs or drugs currently under clinical trials (Figure 2).
Subclonal analysis
Compared to traditional serological markers, ctDNA directly responds to tumor burden and may be used as a potential marker for tumor monitoring. PyClone (26) was used to infer the subclonal architecture of plasma samples before and after treatment. Figure 3 shows the clone number and the mean allele frequency (AF) of the major clone of the plasma samples before treatment. On average, each sample carries 3.56 clones (2-10). The clone number and the mean AF of the major clone are shown in Figure 3. The results suggested that the clone number and the mean AF of the major clone were positively correlated. However, more samples are needed to test this assumption.

To investigate the efficacy of ctDNA in tumor treatment monitoring in liver cancer patients, the mutation frequency mean value of the major clone in plasma ctDNA was used to reflect the overall ctDNA level in the present study. PyClone was used to infer the cancer cell fraction (CCF) in each ctDNA as described previously (27). Clonal was defined as the cluster with the greatest mean CCF variants, and the remaining clusters were considered subclonal (26).
Thirty-four patients with liver cancer were included in this study. Five patients were removed due to lack of AFP concentration information, and 29 patients were included in the following analysis. The AFP concentration in 14 patients was within the normal range (<20 ng/mL) before and after treatment. The AFP concentration in these patients was not suitable as a marker for treatment monitoring. Figure S1 shows the ctDNA levels of these patients, which may be used as a surrogate for AFP. Fifteen patients had an abnormal range of AFP concentrations (>20 ng/mL). Dynamic changes were detected in plasma samples before and after treatment (Figure 4), and the change in AFP concentration was transformed to its percentage change. In general, the ctDNA level was well correlated with AFP concentration at different time points (Figure 4), suggesting that the clinical course of liver cancer can be monitored using ctDNA.

Discussion
In the past few years, the incidence and mortality trend of patients with liver cancer has been significantly decreased in China (28), possibly due to the prevention of hepatitis B virus (HBV) infection through vaccination of infants. However, liver cancer is still one of the most common causes of cancer-associated mortalities in China (28). Poor prognosis in liver cancer patients is common, which may be due to an advanced stage at first diagnosis. Because the survival rates of cancer patients rapidly decline with delayed salvage surgery and late-stage diagnosis (29,30), timely detection is pivotal. Accurate monitoring of treatment response also helps to determine the benefit of new therapeutics and to avoid unnecessary ineffective therapies. Clinically, serial imaging is generally used to assess treatment response; however, radiographic measurements often fail to detect small changes in the early stage of treatment. In clinical management, AFP, CEA, PSA, and CA15-3 are also commonly used to reflect treatment responses (31,32). He et al. (32) showed that the sensitivity of the combined use of markers greatly increases in the diagnosis of gastric cancer. However, in patients with normal concentrations of plasma protein biomarkers in a real clinical setting, the application of these plasma protein biomarkers is limited. In recent years, ctDNA has been regarded as a promising surrogate.
Because the detection of ctDNA is noninvasive, the use of ctDNA to assess treatment response largely avoids the need for repeated invasive biopsy procedures. However, the use of ctDNA to study the extent of clonal heterogeneity in plasma samples before and after treatment is extremely limited. In the present study, we investigated putative clonal clusters and inferred the cellular prevalence in plasma samples of 29 liver patients using PyClone. The results showed that clonal population structures and subclonal dynamics were reflected and captured using ctDNA.
In this study, clonal changes before and after surgery were investigated in 29 patients. These patients were grouped according to the concentration of AFP as follows: one group (group 1, 14 patients) had a normal AFP concentration (Figure S1); and the other group (group 2, 15 patients) had an abnormal AFP concentration (Figure 4). Because the AFP concentration was not suitable as a marker for treatment monitoring for the patients in group 1, the ctDNA levels of these patients could be used as surrogates for AFP for treatment monitoring. The outcomes of patients LZ07, LZ12, LZ20, LZ21, LZ23, LZ27, LZ29, and LZ34 were successfully followed up. Patients LZ21, LZ27, LZ29, and LZ34 had significantly lower ctDNA levels after treatment (Figure S1), suggesting a better prognosis after surgery. Good prognosis was observed in these 4 patients, and no evidence of recurrence was found. Patients LZ12 and LZ23 had elevated ctDNA levels after treatment (Figure S1), suggesting residual tumor formation or recurrence after surgery. Recurrences were observed in April and June in 2021 for patients LZ12 and LZ23, respectively. We also observed a contradictory association between ctDNA levels and clinical outcome in patients LZ07 and LZ20 (Figure S1). Patient LZ07 had significantly lower ctDNA levels after treatment, suggesting better prognosis; however, patient LZ07 died shortly after surgery. In contrast, patient LZ20 had elevated ctDNA levels after treatment; however, good prognosis was observed in patient LZ20. We failed to obtain any further information for patients LZ01, LZ06, LZ10, LZ18, LZ31 and LZ32 due to loss of contact information. For patients with poor AFP sensitivity, these results suggested that ctDNA detection has great potential in monitoring therapeutic effects.
In most cases (12/15; LZ03, LZ05, LZ08, LZ13, LZ15, LZ17, LZ19, LZ22, LZ24, LZ25, LZ26, and LZ28) in group 2, the ctDNA levels were generally correlated well with the changes in plasma samples on computerized AFP concentration. However, some patients had large fluctuations in AFP concentration (Figure 4). Combined with clinical information, the ctDNA levels were more consistent with the clinical condition of the patients, suggesting that ctDNA may be more suitable for monitoring the therapeutic effects in liver cancer patients. We also identified different changes between ctDNA levels and AFP concentration in some patients (such as patients LZ04, LZ11, and LZ30). We further analyzed the clinical information of these patients. The ctDNA level of the plasma sample of patient LZ04 was elevated temporarily at 0.5 months after surgery and decreased at 1.5 months after surgery, which may be related to the insufficient sampling time point of the patient. Patient LZ11 also had insufficient sampling time points. The ctDNA level of the plasma sample of patient LZ30 was elevated at 0.3 months after surgery, which reflected the detection of residual disease using ctDNA. The CT result of this patient in January 2018 showed lipiodol accumulation and uneven density of the right posterior lobe of the liver, which indicated the recurrence of the tumor after surgery. The AFP concentration of the patient increased greatly in May 2018, which also reflected the recurrence of the disease. For patients LZ15 and LZ22, the ctDNA levels after treatment were also decreased, which was consistent with the AFP concentrations. The unusual trends for patients LZ15 and LZ22 shown in Figure 4 may be due to the sampling time points of AFP and ctDNA being different in these two samples. Together, these results suggested that ctDNA can serve as a highly sensitive and real-time marker for monitoring liver cancer and the prognosis of liver cancer. However, more samples and further analysis of the treatment response are still needed.
There were several limitations in this study. Due to the lack of tumor tissue samples of the patients, we were unable to assess the concordance of somatic mutations between tumor DNA and ctDNA in our cohort. Some reports have demonstrated the relationship between ctDNA level and progression-free survival (PFS) and OS (33). Considering the high survival rate of the patients at the time of submission of this paper, the investigation of the association of PFS and OS with ctDNA levels may be another ingesting topic in the future.
Conclusions
In summary, we used PyClone to explore the subclonal architecture and cancer cell fraction in all plasma samples. To the best of our knowledge, this is the first study employing ctDNA in liver cancer for subclonal analysis using the BGISEQ-500 platform. The results showed that ctDNA can be used in the monitoring of liver cancer patients.
Acknowledgments
We thank all the blood donors for their invaluable contribution to this study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tcr-21-1005
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-1005
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1005). Yan Sun reports that this work was supported by the National Natural Science Foundation of China (81502593 to Yan Sun). Jing Yu reports that this work was in part supported by Shenzhen Engineering Laboratory for Innovative Molecular Diagnostics {DRC-SZ[2016]884}. Except Xiaoyu Kong and Xiaolin Zheng, all authors report they are from BGI-Shenzhen.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of BGI (BGI-IRB 15136). Informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol 2014;25:2304-13. [Crossref] [PubMed]
- MANDEL P. METAIS P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Shapiro B, Chakrabarty M, Cohn EM, et al. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 1983;51:2116-20. [Crossref] [PubMed]
- Trevisan França de Lima L, Broszczak D, Zhang X, et al. The use of minimally invasive biomarkers for the diagnosis and prognosis of hepatocellular carcinoma. Biochim Biophys Acta Rev Cancer 2020;1874:188451. [Crossref] [PubMed]
- Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010;2:20ra14. [Crossref] [PubMed]
- van der Vaart M, Semenov DV, Kuligina EV, et al. Characterisation of circulating DNA by parallel tagged sequencing on the 454 platform. Clin Chim Acta 2009;409:21-7. [Crossref] [PubMed]
- Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Chen JG, Zhang SW. Liver cancer epidemic in China: past, present and future. Semin Cancer Biol 2011;21:59-69. [Crossref] [PubMed]
- Mäbert K, Cojoc M, Peitzsch C, et al. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol 2014;90:659-77. [Crossref] [PubMed]
- Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res 2013;73:6384-8. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267-73. [Crossref] [PubMed]
- Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet 2012;44:1117-21. [Crossref] [PubMed]
- Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8. [Crossref] [PubMed]
- Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760-4. [Crossref] [PubMed]
- Li M, Zhao H, Zhang X, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 2011;43:828-9. [Crossref] [PubMed]
- Kawai-Kitahata F, Asahina Y, Tanaka S, et al. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J Gastroenterol 2016;51:473-86. [Crossref] [PubMed]
- Woo HG, Kim SS, Cho H, et al. Profiling of exome mutations associated with progression of HBV-related hepatocellular carcinoma. PLoS One 2014;9:e115152. [Crossref] [PubMed]
- Jhunjhunwala S, Jiang Z, Stawiski EW, et al. Diverse modes of genomic alteration in hepatocellular carcinoma. Genome Biol 2014;15:436. [PubMed]
- Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422-33. [Crossref] [PubMed]
- Kalinina O, Marchio A, Urbanskii AI, et al. Somatic changes in primary liver cancer in Russia: a pilot study. Mutat Res 2013;755:90-9. [Crossref] [PubMed]
- Cleary SP, Jeck WR, Zhao X, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693-702. [Crossref] [PubMed]
- Bratman SV, Newman AM, Alizadeh AA, et al. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn 2015;15:715-9. [Crossref] [PubMed]
- Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396-8. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Gleber-Netto FO, Braakhuis BJ, Triantafyllou A, et al. Molecular events in relapsed oral squamous cell carcinoma: Recurrence vs. secondary primary tumor. Oral Oncol 2015;51:738-44. [Crossref] [PubMed]
- Yom SS, Machtay M, Biel MA, et al. Survival impact of planned restaging and early surgical salvage following definitive chemoradiation for locally advanced squamous cell carcinomas of the oropharynx and hypopharynx. Am J Clin Oncol 2005;28:385-92. [Crossref] [PubMed]
- Tachibana M, Takemoto Y, Nakashima Y, et al. Serum carcinoembryonic antigen as a prognostic factor in resectable gastric cancer. J Am Coll Surg 1998;187:64-8. [Crossref] [PubMed]
- He CZ, Zhang KH, Li Q, et al. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol 2013;13:87. [Crossref] [PubMed]
- Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun 2018;9:3114. [Crossref] [PubMed]