
Down-regulation of PSMD4 can attenuate autophagy, enhance the accumulation of intracellular ROS, and increase the sensitivity of epithelial ovarian cancer to carboplatin by inhibiting the NF-κB pathway
Introduction
Ovarian cancer is a common tumor with an incidence of about 20–30% of female genital tract malignancies (1). Global Cancer Statistics 2020 estimated that there were 295,414 new cases of ovarian cancer and 184,799 deaths from ovarian cancer (2). Studies have shown that about 70% of ovarian cancer patients are already in the advanced stage (FIGO stage III and IV) at the time of diagnosis, and the 5-year survival rate of advanced patients is only about 29% (3).
Epithelial ovarian cancer (EOC) is the most common type of ovarian cancer, accounting for about 90% of new cases (4). At present, the standard clinical treatment plan is based on surgery supplemented with platinum-based combined chemotherapy, but the treatment effect has not been significantly improved. Currently, the first-line chemotherapy drug for EOC is carboplatin (5). Despite the initial positive response to chemotherapy, many patients gradually developed drug resistance and disease recurrence, and eventually became resistant to platinum, which made chemotherapy for EOC ineffective. Therefore, finding the key molecules that affect the sensitivity of carboplatin in EOC may help predict the therapeutic effect and better adjust the treatment plan. At the same time, this may help the development of carboplatin sensitizers or combination therapy drugs, and enhance the efficacy of carboplatin in the treatment of EOC.
Autophagy is a physiological process that is ubiquitous in normal cells and EOC cells (6). When the cells become cancerous, the level of autophagy increases under the stimulation of various pathological factors such as hypoxia, starvation, radiotherapy, and chemotherapy (7,8). Its protective effect makes the cells better adapt to the harsh living environment to maintain survival, promote tumor progression, and produce multi-drug resistance. After chemotherapy drugs act on cancer cells, they induce protective autophagy and inhibit apoptosis. A study has pointed out that a certain level of autophagy persists in the EOC cell line A2780, and low-dose carboplatin can up-regulate the expression of microtubule-associated protein 1 light chain 3 (LC3)-II (9).
Proteasome 26S subunit, non-ATPase 4 (PSMD4) is a regulatory subunit of the 19S regulatory component of the 26S proteasome (10). The PSMD4 structure contains ubiquitin-protein action motifs, which interact with ubiquitinated proteins to mediate the degradation of ubiquitinated proteins (11). In the process of tumor occurrence and development, the protein metabolism degradation pathway is often disturbed, and the intracellular homeostasis is disrupted. A previous study has found that PSMD4 may be related to the poor prognosis of patients with multiple myeloma and resistance to bortezomib, and they also found that the low expression of PSMD4 significantly enhanced the killing effect of bortezomib on tumor cells (12). However, the specific role of PSMD4 in the carboplatin sensitivity of EOC is still unclear.
In this study, based on the second-generation sequencing technology and bioinformatics methods, we first discovered that PSMD4 is the core molecule in the carboplatin resistance regulatory network of EOC, and its expression is up-regulated in carboplatin-resistant cells. In vitro and in vivo experiments have shown that the down-regulated expression of PSMD4 is closely related to the increase in carboplatin sensitivity of EOC. Further mechanism studies have found that inhibiting the expression of PSMD4 may inhibit the activation of the nuclear factor kappa-B (NF-κB) pathway, thereby inhibiting autophagy, promoting carboplatin-induced reactive oxygen species (ROS) accumulation, and ultimately enhancing the sensitivity of EOC cells to carboplatin.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1389).
Methods
Cell culture and transfection
Human EOC cell lines [TOV-21G (ATCC Cat# CRL-11730, RRID:CVCL_3613), SK-OV-3 (ATCC Cat# HTB-77, RRID:CVCL_0532), TOV-112D (ATCC Cat# CRL-11731, RRID:CVCL_3612), ES-2 (ATCC Cat# CRL-1978, RRID:CVCL_3509), and OVCAR-3 (CLS Cat# 300307/p690_NIH:Ovcar-3, RRID:CVCL_0465)] were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). TOV-21G, SK-OV-3, TOV-112D, ES-2, and OVCAR-3 cells were cultured in RPMI 1640 Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin at 37 °C in 5% CO2.
The EOC cells in the logarithmic growth phase were seeded into a six-well plate at a density of 5×105 cells/well, and cultured in a complete culture medium without antibiotics. To further study the functional role of PSMD4 in EOC cells, we established a PSMD4 silencing or overexpression model in EOC cells. A PSMD4 knockdown short hairpin RNA (shRNA) construct (shPSMD4) was purchased from Genechem (China), and the non-targeting shRNA vector was used as a negative control. The PSMD4 overexpression vector (OV-PSMD4) was purchased from Gemma Pharmaceutical Technology Co., Ltd. (China), and the empty vectors were used as a negative control. EOC cells were cultured in 60 mm cell culture plates and grown to 50–70% confluence and the plasmids were separately transfected into EOC cells. Transfection was conducted using Neofect DNA transfection reagent (Neofect Biotech Co., Ltd., China), which was used according to the manufacturer’s protocol.
Establishment of carboplatin-resistant cell lines
Carboplatin-resistant cell lines (SK-OV-3/carboplatin and TOV-21G/carboplatin) were established by exposing SK-OV-3 and TOV-21G cells to gradually increasing the dose of carboplatin (0.1–2.0 μg/mL) for 5 months. Two weeks after the withdrawal of the carboplatin treatment, the biological characteristics of the resistant strains were observed. One month after the withdrawal of the carboplatin, the carboplatin-resistant cell lines can still maintain drug resistance. Compared with the half-maximal inhibitory concentration (IC50) when the carboplatin was withdrawn, there was no statistical difference in the IC50 change at 1 month after the carboplatin was withdrawn (P>0.05), indicating that the carboplatin-resistant cell lines have better drug resistance stability.
Second-generation sequencing technology
To screen for genes related to carboplatin resistance in EOC, we performed second-generation sequencing analysis on carboplatin-sensitive and resistant cell lines. The original image data file obtained by Illumina HiseqTM is converted into the original sequencing sequence, and the quality value of the original data is statistically and visually evaluated. We used the DEGSeq or DESeq R function package to standardize the counts of the sample genes and calculate the multiples of gene expression differences. Then, we used the negative binomial distribution test to test the significance of the number of reads. |logFC| >1 and adjust P<0.05 are considered to be significantly differentially expressed genes, and the ggplot2 package is used to draw a volcano map reflecting the overall distribution of differentially expressed genes.
Bioinformatics analysis
This study used Gene Expression Omnibus (GEO) database, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), STRING (https://string-db.org/) database, and DAVID (http://david.abcc.ncifcrf.gov/) database.
The differentially expressed genes related to carboplatin resistance in EOC were entered into the STRING database to search the interaction relationship between the proteins online, and the combined score >0.4 was used as the screening condition. The results obtained are imported into Cytoscape software for visualization processing, and a protein-protein interaction (PPI) network is constructed. Further, we used the DAVID online tool to perform GO function annotation analysis and KEGG pathway enrichment analysis on the selected differentially expressed genes and used the ClusterProfiler function package for visual display.
Finally, through the NCBI-GEO data set (https://www.ncbi.nlm.nih.gov/gds/advanced), the EOC tissue expression profile chip was searched. The GSE33482 EOC tissue chip was selected using “ovarian cancer” AND “carboplatin” as the search keywords. The chip included 21 patients with locally advanced EOC who received carboplatin-based chemotherapy. Patients were finally clinically evaluated 6 months after the end of treatment. Among them, 12 patients were in complete remission and 9 patients were in incomplete remission. We used the limma R package to analyze the differences in the expression profiles of the two groups of patients, and screened out the differential genes related to carboplatin resistance in EOC. In addition, we used the ggplot2, pheatmap, and pROC function packages to draw volcano maps, heat maps, and violin maps to verify the expression of core genes in carboplatin-resistant tissues in EOC.
Transmission electron microscopy
Cells were fixed with 2.5% glutaraldehyde in phosphate buffer. The specimens were post-fixed in 1% Osmium Tetroxide with 0.1% potassium ferricyanide, dehydrated through a graded series of ethanol (30–90%). Images were taken on a HITACHI HT7700 transmission electron microscopy system (HITACHI, Japan) at 80 kV.
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay
Digest the cells in the logarithmic growth phase with trypsin, resuspend the culture medium, and spread a 96-well plate at a cell density of 5×103 cells/well. When the cell confluence reaches 80%, different concentrations of carboplatin are added. After 24 h, discard the drug-containing culture medium, add 20 μL MTS (Shanghai Yihui Biological Technology Co., Ltd., Shanghai, China) to each well, incubate at 37 °C in the dark for 2 to 4 h, and detect each well with a microplate reader. The optical density (OD) was detected at a wavelength of 490 nm. Cell survival rate % = (average value of the experimental group − average value of background plate/average value of control group-average value of background plate) × 100%. All experiments were repeated 3 times. The equation of IC50 of carboplatin was [viability% = 100/(1 + 10[carboplatin concentration]log IC50)].
Quantitative real-time PCR (qRT-PCR) analyses
Total RNA of cell lines was extracted. GAPDH was used as the housekeeping gene. The primer sequences used for qRT-PCR were as follows: 5'-GATCGCATGCTGCCAGCCCAAC-3' (forward) and 5'-GCGCACTTCAGGCCAGCGTCAGA-3' (reverse) for PSMD4; and 5'-CTCGGCTCGACGAGCACA-3' (forward) and 5'-AACGCTGGACGACTGCCCGT-3' (reverse) for GAPDH. The relative level was calculated by the 2−ΔΔCt method.
Western blot
Total protein was extracted separately from EOC cells. A separation gel and concentrated gel were prepared, and the proteins were separated by electrophoresis and then transferred to membranes. The membranes were blocked with 5% skim milk and incubated with antibodies against PSMD4 (1:2,000, GenWay Biotech Inc. Cat# GWB-F0AEDC, RRID:AB_10268465), LC3 (1:1,000, MBL International Cat# PD014, RRID:AB_843283), Beclin-1 (1:1,000, US Biological Cat# B0981-23T, RRID:AB_2064477), P62 (1:2,000, PeproTech Cat# 500-P62-50 μg, RRID:AB_147647), cleaved caspase-3 (1:3,000, Cell Signaling Technology Cat# 8202, RRID:AB_1658166), Bcl-2 (1:2,000, LSBio (LifeSpan) Cat# LS-C438-1000, RRID:AB_1272355), Bax (1:2,000, GenWay Biotech Inc. Cat# 18-661-15197-0.1 mg, RRID:AB_516294), NF-κB p65 (1:2,000; Cell Signaling Technology Cat# 3034, RRID:AB_330561), IkappaB-alpha (IκBα, 1:1,000, ABclonal Cat# A19714, RRID:AB_2862749), Lamin B (1:2,000, LSBio (LifeSpan) Cat# LS-B2005-50, RRID:AB_1275098), and β-actin (1:3,000, Miltenyi Biotec Cat# 130-120-277, RRID:AB_2857517). Finally, the secondary antibody was incubated for 2 hours, and the gray band of the protein band was detected by chemiluminescence.
Flow cytometry apoptosis detection
Cell apoptosis was determined by flow cytometry after staining with Annexin V (FITC-conjugated) apoptosis kit. Briefly, EOC cells were stained with 5 µL Annexin 5-FITC and propidium iodide (PI), respectively, and incubated for 15 min in the dark, and then analyzed by flow cytometry.
Hoechst staining
EOC cells were incubated with 5-ethynyl-20-deoxyuridine (EdU, Shanghai Huicheng Biological Technology Co., Ltd., Shanghai, China) for 5 hours. After three washes with phosphate buffer saline, cells were treated with a 1× Apollo® reaction cocktail (100 μL/well) for 30 min. After that, each well was stained with Hoechst 33342 (5 μg/mL, 100 µL/well) for 30 min.
Intracellular ROS detection
First, dilute 2,7-Dichlorodi-hydrofluorescein diacetate (DCFH-DA, Beijing Biolab Technology Co., Ltd., China) with serum-free culture medium at 1:1,000 to make the final concentration 10 µmol/L. After harvesting the cells in a centrifuge tube, suspend them in the diluted DCFH-DA at a cell concentration of 1×106–2×107/mL, and incubate in a cell culture incubator at 37 °C for 20 minutes. During this period, mix upside down once at an interval of 3 to 5 minutes so that the cells and the probe are in full contact. Wash the cells three times with a serum-free medium to fully remove the DCFH-DA that has not entered the cells. Transfer the stained cells to a flow tube for testing.
Immunohistochemistry
Immunohistochemistry staining was performed on paraffin sections with rabbit monoclonal antibodies directed against PSMD4 (1:100; Enzo Life Sciences Cat# PAB0242, RRID:AB_1532684), NF-κB p65 (1:100; Cell Signaling Technology Cat# 3034, RRID:AB_330561), and IκBα (1:100, ABclonal Cat# A19714, RRID:AB_2862749). The biotinylated goat anti-rabbit secondary antibody (1:5,000, Signalway Cat# L3012, RRID:AB_895483) was incubated at 4 °C. Finally, the sections were incubated with DAB substrate for 5 min. To validate the PSMD4, NF-κB p65, and IκBα antibodies used in immunohistochemistry experiments, liver cancer and para-cancerous tissue samples were used as positive and negative controls (Figure S1).
In vivo tumor xenograft
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Humane care was given during the experimental animal breeding and experimental procedures following the 3R principle of experimental animals. Experiments were performed under a project license (No. K2019001A011) granted by the Ethics Committee of the Kunshan Second People’s Hospital, in compliance with the Kunshan Second People’s Hospital’s guidelines for the care and use of animals.
Twelve female BALB/c nude mice (20–22 g, 4–6 weeks old) were obtained from Beijing Weitong Lihua (Beijing, China). The culture of mice and subsequent experiments were carried out in SPF animal laboratories. The rearing environment of the mice is 12 h light and 12 h dark alternately, and the mice freely eat water and food. After 1 week of adaptive cultivation, follow-up experiments were carried out.
Twelve mice were divided into four groups (SK-OV-3/carboplatin group and TOV-21G/carboplatin group) according to the random number table method, three in each group. Collect shPSMD4-SK-OV-3/carboplatin, shPSMD4-TOV-21G/carboplatin, negative control SK-OV-3/carboplatin, and TOV-21G/carboplatin cells in a logarithmic growth phase. After trypsin digestion, wash with PBS solution by centrifugation, prepare cell suspension with PBS, adjust the cell density to 2×104 cells/μL after counting, and mix with Matrigel glue at a ratio of 1:1 before use. Then, cells were subcutaneously injected into nude mice to establish the xenograft model. After the model was successfully established, carboplatin injection was used to inject the nude mice into the abdominal cavity once every 3 days at a dose of 3 mg/kg for a total of 5 times. The effect of carboplatin on transplanted tumors in mice was observed. The diameter of each xenograft tumor was measured before each administration. The tumor volume was calculated regularly according to the formula: volume of tumor (mm3) = (length × width2)/2. The nude mice were sacrificed 3 days after the last dose.
Statistical analysis
The data analysis and processing software used in this study include R version 3.6.1 (81 megabytes, 32/64 bit), Cytoscape version 3.7.2 (http://www.cytoscape.org/), GraphPad Prism version 7.0 (GraphPadSoftware, La Jolla, CA, USA). All experiments were performed in triplicate unless specified. Results were represented as the mean ± standard deviation. The differences between normally distributed numeric variables were evaluated by Student’s t-test. One-way analysis of variance (ANOVA) was used for the comparison among multiple groups if the variance was homogeneous. Multiple comparisons between the groups were performed using the S-N-K method. Correlations were analyzed using the Pearson correlation analysis. P<0.05 was considered significant.
Results
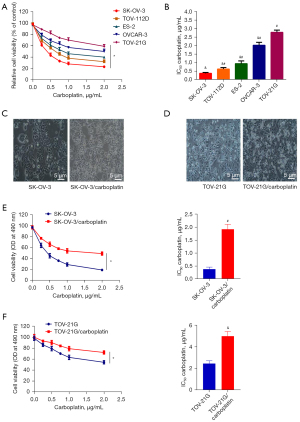
Characterization of carboplatin resistance in SK-OV-3/carboplatin and TOV-21G/carboplatin cells
Carboplatin cytotoxicity for TOV-21G, SK-OV-3, TOV-112D, ES-2, and OVCAR-3 cell lines was examined. SK-OV-3 cells possessed the most sensitivity to carboplatin accompanied by the lowest IC50, whereas TOV-21G cells were found to be the least sensitive accompanied by the highest IC50 (Figure 1A,1B). Then, the carboplatin-resistant cell lines (SK-OV-3/carboplatin and TOV-21G/carboplatin) were established. Observing the cell morphology of each group under a light microscope, it can be seen that SK-OV-3/carboplatin and TOV-21G/carboplatin cells are more slender, with binuclear cells visible, the adhesion between cells disappeared, and there are obvious intracellular blanks (Figure 1C,1D). Furthermore, we characterized carboplatin resistance in SK-OV-3/carboplatin and TOV-21G/carboplatin cells. After exposure to gradually increasing dose of carboplatin (0.1–2.0 μg/mL) for 2 days, the IC50 values were 1.87±0.19 μg/mL and 4.89±0.54 μg/mL for SK-OV-3/carboplatin and TOV-21G/carboplatin cells, respectively, showing a 4.8- and 1.8-fold increase as compared to parental cell lines (SK-OV-3 cells: 0.39±0.08 μg/mL, Figure 1E; TOV-21G cells: 2.68±0.47 μg/mL, Figure 1F).
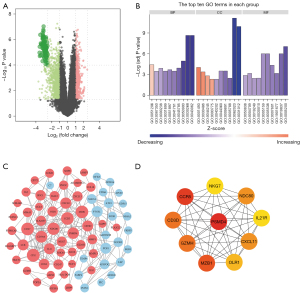
Screening and analysis of carboplatin resistance-related genes in EOC
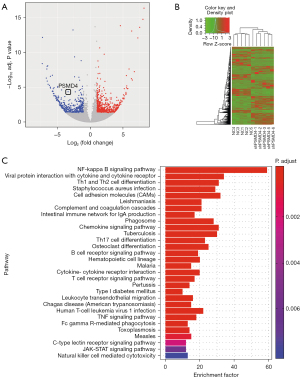
We then chose SK-OV-3 and SK-OV-3/carboplatin cell lines for sequencing analysis, and a total of 526 differentially expressed mRNAs, including 389 up-regulated genes and 137 down-regulated genes, were found (Figure 2A). Subsequently, we performed GO enrichment analysis on the differentially expressed genes related to carboplatin resistance. The enrichment of the top 10 GO projects is shown in Figure 2B. In the biological process, it is mainly concentrated in inflammation. In the cell components, it is mainly enriched in the adhesion and connection between cells. In terms of molecular function, it is mainly enriched in protein kinase activity. Then, we searched the interactions between 526 differentially expressed genes related to EOC carboplatin resistance through the STRING database and constructed a PPI network (Figure 2C). Finally, we screened the 10 differentially expressed genes (PSMD4, CCR5, NKG7, NDC80, IL21R, MZB1, OLR1, GZMH, CD3D, CXCL11) with the highest degree as core genes (Figure 2D).
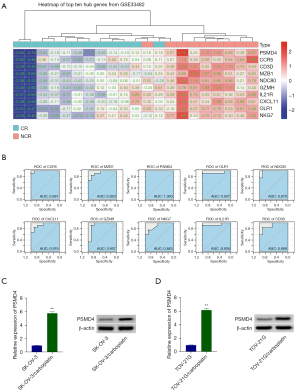
We screened the array data set GSE33482 from the NCBI-GEO database for mRNAs difference analysis. This data set contains tissue samples from EOC carboplatin-resistant and sensitive patients. Subsequently, the pheatmap package was used to perform a bi-clustering analysis on the 10 core genes in the PPI network, and the results are shown in Figure 3A. In addition, we also used the pROC package to perform receiver operating characteristic (ROC) curve analysis on 10 core genes in the PPI network. PSMD4 possesses a high prognostic value in EOC carboplatin-resistant patients [area under the curve (AUC) =1.0, Figure 3B]. Based on the above results, we used qRT-PCR and Western blot to detect the expression of PSMD4 in SK-OV-3, SK-OV-3/carboplatin, TOV-21G, and TOV-21G/carboplatin cells, respectively. We found that compared with SK-OV-3 and TOV-21G cells, the mRNA and protein levels of PSMD4 in SK-OV-3/carboplatin (P<0.01, Figure 3C) and TOV-21G/carboplatin (P<0.01, Figure 3D) cells were significantly increased. All these results are consistent with the results of our previous bioinformatics analysis.
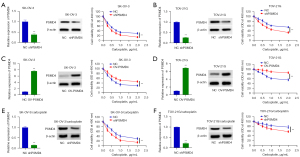
Inhibiting PSMD4 can promote the sensitivity of carboplatin-resistant EOC cells to carboplatin in vitro and in vivo
To further verify the relationship between PSMD4 expression and EOC carboplatin sensitivity, SK-OV-3, SK-OV-3/carboplatin, TOV-21G, and TOV-21G/carboplatin cells were transfected with OV-PSMD4, shPSMD4, and NC. Different concentrations of carboplatin were given to observe the changes in cell viability. QRT-PCR and Western blot results found that the expression level of PSMD4 in EOC cells transfected with shPSMD4 plasmid was significantly lower than that of the control group (P<0.01, Figure 4A,4B). The expression level in EOC cells transfected with PSMD4 overexpression plasmid was significantly higher than that in the control group (P<0.01, Figure 4C,4D). Compared with the control group, the cells in the PSMD4 low expression group were significantly more sensitive to carboplatin (P<0.01, Figure 4A,4B), while the PSMD4 overexpression group cells were significantly less sensitive to carboplatin (P<0.01, Figure 4C,4D). To investigate the specific function of PSMD4 in SK-OV-3/carboplatin and TOV-21G/carboplatin cells, we achieved PSMD4 silencing by infecting cells with shPSMD4. Similarly, consistent results can be observed in the SK-OV-3/carboplatin and TOV-21G/carboplatin cell lines (P<0.01, Figure 4E,4F).
The killing effect of carboplatin is cell apoptosis induced by DNA damage (13). Therefore, to further explore the effect of PSMD4 on carboplatin sensitivity of EOC cells, flow cytometry was performed. We found that PSMD4 knockdown enhanced cell apoptosis (Figure 5A,5B), and nuclear fragmentation (Figure 5C) in SK-OV-3/carboplatin and TOV-21G/carboplatin cells. Moreover, PSMD4 knockdown inhibited autophagy in SK-OV-3/carboplatin and TOV-21G/carboplatin cells (Figure 5D). In vivo experiments further confirmed the results of cell experiments. The anti-tumor effect of carboplatin on PSMD4 down-regulated xenograft mice was significantly better than the control group, indicating that nude mice with low PSMD4 expression are more sensitive to carboplatin (Figure 5E-5G).
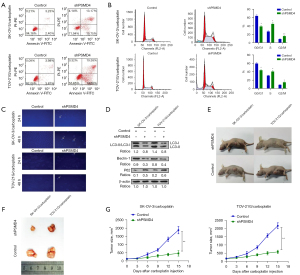
Screening and enrichment of differentially expressed genes in shPSMD4-SK-OV-3/carboplatin and NC-SK-OV-3/carboplatin cells
Then, we sequenced shPSMD4-SK-OV-3/carboplatin and NC-SK-OV-3/carboplatin and found a total of 869 differentially expressed mRNAs (P<0.05 and |LogFC| >1), of which 254 upregulated genes and 615 down-regulated genes (Figure 6A,6B). KEGG enrichment analysis was used to explore the differentially expressed genes in shPSMD4-SK-OV-3/carboplatin cells. The results showed that the KEGG pathway was mainly enriched in the NF-κB signaling pathway (Figure 6C). Combined with previous studies (14,15), we believe that there is a close regulatory relationship between PSMD4 and NF-κB. We speculate that PSMD4 may partly mediate the NF-κB signaling pathway and affect EOC carboplatin sensitivity.
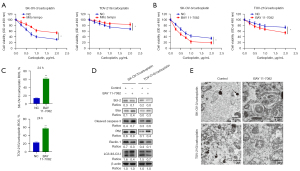
Inhibition of the NF-κB pathway can weaken autophagy of carboplatin-resistant EOC cells, increase intracellular ROS levels, and enhance carboplatin sensitivity
Studies have confirmed that carboplatin can induce the accumulation of ROS, and the accumulation of ROS is an important part of cell apoptosis (16,17). Mito Tempo is a mitochondrial-targeted antioxidant that can inhibit the generation and accumulation of ROS (18). In this study, we first detected the changes in cell viability of SK-OV-3/carboplatin and TOV-21G/carboplatin cells after the combined usage of Mito Tempo and carboplatin for 24 h. We found that the survival rate of SK-OV-3/carboplatin and TOV-21G/carboplatin cells added to Mito Tempo was significantly higher than that of the control group (Figure 7A). Subsequently, we combined the NF-κB inhibitor (BAY 11-7082) and carboplatin to treat SK-OV-3/carboplatin and TOV-21G/carboplatin for 24 h. MTS results showed that the survival rate of SK-OV-3/carboplatin and TOV-21G/carboplatin cells added with NF-κB inhibitor was significantly lower than that of the control group (Figure 7B). Flow cytometry results also indicate that NF-κB inhibitors can significantly up-regulate the ROS content and induce cell apoptosis under the action of carboplatin in SK-OV-3/carboplatin and TOV-21G/carboplatin cells (Figure 7C). Western blot results showed that NF-κB inhibitors can promote apoptosis and inhibit autophagy of SK-OV-3/carboplatin and TOV-21G/carboplatin cells (Figure 7D). Transmission electron microscopy results further confirmed the conclusion (Figure 7E).
Inhibition of PSMD4 expression may enhance carboplatin sensitivity of EOC cells by mediating NF-κB pathway inhibition
Subsequently, we detected the expression of NF-κB p65 and IκBα protein when PSMD4 was inhibited, and observed the influence of the NF-κB pathway by detecting the expression of NF-κB p65 protein in the cytoplasm and nucleus, respectively. The level of NF-κB p65 protein in the cytoplasm of shPSMD4-SK-OV-3/carboplatin and shPSMD4-TOV-21G/carboplatin groups was significantly higher than that of the control group, while the level of NF-κB p65 protein in the nucleus was significantly lower than that of the control group (P<0.05, Figure 8A). At the same time, the expression of IκBα protein in shPSMD4-SK-OV-3/carboplatin and shPSMD4-TOV-21G/carboplatin groups was significantly higher than that in the control group (P<0.05, Figure 8A). Since the NF-κB protein enters the nucleus to perform transcriptional regulation functions, when the NF-κB protein entering the nucleus is significantly reduced, it indicates that the activation of the NF-κB pathway may be inhibited (19). At the same time, IκBα protein is the regulatory protein of the NF-κB classical pathway, and the increase in its expression level also indirectly confirms the inhibition of NF-κB function (19).

To further confirm the influence of PSMD4 on the NF-κB pathway, we performed immunohistochemical detection of PSMD4, NF-κB p65, and IκBα protein expression in EOC nude mice xenograft tumors. The results showed that the positive signal of PSMD4 and NF-κB p65 in the shPSMD4 group was lower than that of the control group, and the positive signal of IκBα was higher than that of the control group (P<0.05, Figure 8B). The above results indicate that the down-regulation of PSMD4 may inhibit the NF-κB p65 and promote the IκBα expression.
Discussion
Ovarian cancer is one of the leading causes of cancer-related deaths in women worldwide (2). For the treatment of ovarian cancer, surgery and radiotherapy are the first and relatively more effective treatments. However, EOC patients in some underdeveloped areas are often diagnosed at a later stage and are no longer suitable for surgery. In addition, due to a large number of patients, the waiting time for the radiotherapy machine is also very long. In this case, carboplatin-based chemotherapy is often selected. It is worth noting that for patients with advanced EOC who have recurrence and metastasis that are not suitable for local treatment, carboplatin-based systemic treatment is often used (20). Carboplatin resistance, whether it is inherent resistance or acquired resistance, will seriously affect the effectiveness of treatment. In response to this situation, we have launched a study on how to improve the sensitivity of carboplatin in EOC.
In this study, we compared SK-OV-3 and SK-OV-3/carboplatin cell lines by transcriptome analysis and comparison. A total of 526 differentially expressed genes were screened, including 389 up-regulated differentially expressed genes and 137 down-regulated differentially expressed genes. After PPI network analysis, GEO tissue microarray data, qRT-PCR, and Western blot results, the PSMD4 protein was finally determined. PSMD4 protein contains ubiquitin protein action motifs, which can mediate the degradation of ubiquitinated proteins. In the process of protein metabolism and degradation, ubiquitin transporters are believed to promote the degradation of ubiquitin proteins (11). However, related studies have shown that the ubiquitin-proteasome system is also inhibited by ubiquitin transporters (21,22). Cell experiments found that the synthesis of ubiquitin chains was inhibited by Rad23, and mouse experiments showed that the degradation of p53 and IκB molecules in cells was inhibited by hPLIC-2, and there was an interaction between PSMD4 and Rad23 and hPLIC-2 (21,22). A recent study has found that the interaction between the proteasome and the ubiquitin-binding protein Dsk2 is affected by PSMD4, thereby acting on the ubiquitin-proteasome degradation system (23). PSMD4 can also be hydrolyzed into small fragments and then secreted out of the cell, participating in a variety of biological processes, suggesting that the role of PSMD4 in tumors is complex (24).
To prove the relationship between PSMD4 expression and carboplatin sensitivity of EOC cells, in this study, we used MTS to detect the survival of PSMD4 low- and over-expressing cells under the action of carboplatin. It was found that the survival rate of PSMD4 down-regulated cells was significantly reduced. We then used flow cytometry and Hoechst staining to compare the apoptosis changes of carboplatin-induced cells with low PSMD4 expression and control cells. It was found that the apoptotic rate of cells with low PSMD4 expression increased significantly, and the results of Western blot also proved that the levels of cleaved caspase-3 and Bax increased significantly, and the expression of Bcl-2 decreased significantly in cells with low PSMD4 expression. At the same time, the results of in vivo animal experiments also showed that the growing volume and rate of PSMD4 down-regulated mouse tumors after carboplatin treatment were significantly lower than those in the control group, and the sensitivity of nude mice with low PSMD4 expression to carboplatin increased. These results indicate that the inhibition of PSMD4 expression can promote carboplatin-induced EOC cell apoptosis and increase the carboplatin sensitivity of EOC cells. The positive effect of the PSMD4 in improving the chemotherapy sensitivity of EOC patients has been confirmed in some studies (12,25,26). What’s more, one study has found that the anti-angiogenic drug cediranib can directly inhibit or inhibit the expression of the PSMD4 gene by inducing hypoxia, and then down-regulate the homologous DNA repair of tumor cells, thereby acting as a DNA repair inhibitor to enhance the sensitivity of chemotherapy drugs (10). However, it is worth noting that we constructed mouse tumor models by subcutaneous injection, which prevented us from observing the effect of down-regulation of PSMD4 on EOC metastasis. Future research can pay more attention to the effect of PSMD4 on intraperitoneal metastasis of EOC.
To further explore the mechanism of inhibiting PSMD4 in enhancing EOC carboplatin sensitivity, we sequenced shPSMD4-SK-OV-3/carboplatin and NC-SK-OV-3/carboplatin cells using second-generation sequencing technology. The results showed that differentially expressed genes were mainly enriched in the NF-κB signaling pathway. A previous study has found that adding carboplatin to cervical cancer SiHa and CaSki cells can activate the NF-κB signaling pathway and induce the expression of NF-κB (27). Our Western blot results also found that in EOC carboplatin-resistant cells, the nuclear expression of NF-κB p65 was significantly increased, and the expression of IκBα was significantly reduced, indicating that the up-regulation of NF-κB may inhibit EOC carboplatin sensitivity. We believe that the continuously activated NF-κB may provide a mechanism to prevent EOC cell apoptosis, which in turn leads to reduced carboplatin sensitivity.
A large number of studies have found that there are many interactions between the NF-κB signaling pathway and ROS (28-30). The transcription of NF-κB-dependent genes can affect the production of ROS in cells. Studies have pointed out that NF-κB inhibits the accumulation of ROS by inducing the expression of downstream genes FHC, thereby preventing apoptosis caused by pro-apoptotic signals (29). In this study, we used mitochondrial targeting antioxidant Mito Tempo combined with carboplatin to act on SK-OV-3/carboplatin and TOV-21G/carboplatin cells. It was found that compared with the control group, Mito Tempo can reduce carboplatin-induced apoptosis, indicating that the sensitivity of EOC cells to carboplatin may be closely related to ROS. Our inference is also supported by previous literature reports (30). At the same time, in the study, we also found that NF-κB inhibitors can enhance carboplatin-induced EOC drug-resistant cells SK-OV-3/carboplatin and TOV-21G/carboplatin apoptosis and increase the content of intracellular ROS. Therefore, we speculate that inhibition of the NF-κB pathway promotes the accumulation of ROS in EOC-resistant cells induced by carboplatin, thereby inducing EOC cell apoptosis.
Conclusions
In summary, the down-regulation of PSMD4 may inhibit the activation of the NF-κB pathway, inhibit autophagy, and up-regulate the level of intracellular ROS accumulation, thereby promoting carboplatin-mediated EOC cell apoptosis and enhancing carboplatin sensitivity. This study may provide a new theoretical basis for the study of the mechanism of EOC carboplatin sensitivity and EOC chemotherapy. At the same time, PSMD4 and NF-κB pathway-related molecules may become prognostic targets for carboplatin therapy and drug resistance reversal targets for EOC patients, which will help prolong the survival of EOC patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1389
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-1389
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-1389
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1389). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. K2019001A011) granted by the Ethics Committee of the Kunshan Second People’s Hospital, in compliance with the Kunshan Second People’s Hospital’s guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lyon KA, Huang JH. Bevacizumab combined with chemotherapy in platinum-resistant ovarian cancer: beyond the AURELIA trial. Transl Cancer Res 2020;9:2164-7. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Palmqvist C, Staf C, Mateoiu C, et al. Increased disease-free and relative survival in advanced ovarian cancer after centralized primary treatment. Gynecol Oncol 2020;159:409-17. [Crossref] [PubMed]
- Amin N, Chaabouni N, George A. Genetic testing for epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol 2020;65:125-38. [Crossref] [PubMed]
- Alvarez Secord A, Bell Burdett K, Owzar K, et al. Predictive blood-based biomarkers in patients with epithelial ovarian cancer treated with carboplatin and paclitaxel with or without bevacizumab: results from GOG-0218. Clin Cancer Res 2020;26:1288-96. [Crossref] [PubMed]
- Gan X, Zhu H, Jiang X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer 2020;19:45. [Crossref] [PubMed]
- Zhao L, Kong X, Zhong W, et al. FTO accelerates ovarian cancer cell growth by promoting proliferation, inhibiting apoptosis, and activating autophagy. Pathol Res Pract 2020;216:153042. [Crossref] [PubMed]
- Chen X, Sun Y, Wang B, et al. Prognostic significance of autophagy-related genes Beclin1 and LC3 in ovarian cancer: a meta-analysis. J Int Med Res 2020;48:300060520968299. [Crossref] [PubMed]
- Ali JL, Lagasse BJ, Minuk AJ, et al. Differential cellular responses induced by dorsomorphin and LDN-193189 in chemotherapy-sensitive and chemotherapy-resistant human epithelial ovarian cancer cells. Int J Cancer 2015;136:E455-69. [Crossref] [PubMed]
- AydoĞan TÜrkoĞlu S. Dayi G, KÖÇkar F. Upregulation of PSMD4 gene by hypoxia in prostate cancer cells. Turk J Biol 2020;44:275-83. [Crossref] [PubMed]
- Fejzo MS, Anderson L, Chen HW, et al. Proteasome ubiquitin receptor PSMD4 is an amplification target in breast cancer and may predict sensitivity to PARPi. Genes Chromosomes Cancer 2017;56:589-97. [Crossref] [PubMed]
- Shaughnessy JD Jr, Qu P, Usmani S, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood 2011;118:3512-24. [Crossref] [PubMed]
- Slootbeek PHJ, Duizer ML, van der Doelen MJ, et al. Impact of DNA damage repair defects and aggressive variant features on response to carboplatin-based chemotherapy in metastatic castration-resistant prostate cancer. Int J Cancer 2021;148:385-95. [Crossref] [PubMed]
- Cheng YM, Lin PL, Wu DW, et al. PSMD4 is a novel therapeutic target in chemoresistant colorectal cancer activated by cytoplasmic localization of Nrf2. Oncotarget 2018;9:26342-52. [Crossref] [PubMed]
- Lin PL, Chang JT, Wu DW, et al. Corrigendum to "Cytoplasmic localization of Nrf2 promotes colorectal cancer with more aggressive tumors via upregulation of PSMD4" Free Radic. Biol. Med. 95 (2016) 121-32. Free Radic Biol Med 2017;104:380-1. [Crossref] [PubMed]
- Stankovic JSK, Selakovic D, Mihailovic V, et al. Antioxidant supplementation in the treatment of neurotoxicity induced by platinum-based chemotherapeutics-a review. Int J Mol Sci 2020;21:7753. [Crossref] [PubMed]
- Mendez N, Alarcón P, Millán C, et al. Vincristine, carboplatin and cisplatin increase oxidative burst induced by PAF in canine neutrophils. Vet Immunol Immunopathol 2020;221:110011. [Crossref] [PubMed]
- Schomberg J, Wang Z, Farhat A, et al. Luteolin inhibits melanoma growth in vitro and in vivo via regulating ECM and oncogenic pathways but not ROS. Biochem Pharmacol 2020;177:114025. [Crossref] [PubMed]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol 2002;2:725-34. [Crossref] [PubMed]
- Machida H, Tokunaga H, Matsuo K, et al. Survival outcome and perioperative complication related to neoadjuvant chemotherapy with carboplatin and paclitaxel for advanced ovarian cancer: a systematic review and meta-analysis. Eur J Surg Oncol 2020;46:868-75. [Crossref] [PubMed]
- Liang RY, Chen L, Ko BT, et al. Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. J Mol Biol 2014;426:4049-60. [Crossref] [PubMed]
- Walters KJ, Kleijnen MF, Goh AM, et al. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 2002;41:1767-77. [Crossref] [PubMed]
- Chen X, Ebelle DL, Wright BJ, et al. Structure of hRpn10 bound to UBQLN2 UBL illustrates basis for complementarity between shuttle factors and substrates at the proteasome. J Mol Biol 2019;431:939-55. [Crossref] [PubMed]
- Cai MJ, Cui Y, Fang M, et al. Inhibition of PSMD4 blocks the tumorigenesis of hepatocellular carcinoma. Gene 2019;702:66-74. [Crossref] [PubMed]
- Misiewicz-Krzeminska I, de Ramón C, Corchete LA, et al. Quantitative expression of Ikaros, IRF4, and PSMD10 proteins predicts survival in VRD-treated patients with multiple myeloma. Blood Adv 2020;4:6023-33. [Crossref] [PubMed]
- Xu H, Van der Jeught K, Zhou Z, et al. Atractylenolide I enhances responsiveness to immune checkpoint blockade therapy by activating tumor antigen presentation. J Clin Invest 2021;131:146832. [Crossref] [PubMed]
- Bravo-Cuellar A, Ortiz-Lazareno PC, Sierra-Díaz E, et al. Pentoxifylline sensitizes cisplatin-resistant human cervical cancer cells to cisplatin treatment: involvement of mitochondrial and NF-kappa B pathways. Front Oncol 2020;10:592706. [Crossref] [PubMed]
- Ramos-Tovar E, Muriel P. Molecular mechanisms that link oxidative stress, inflammation, and fibrosis in the liver. Antioxidants (Basel) 2020;9:1279. [Crossref] [PubMed]
- Kou X, Jing Y, Deng W, et al. Tumor necrosis factor-α attenuates starvation-induced apoptosis through upregulation of ferritin heavy chain in hepatocellular carcinoma cells. BMC Cancer 2013;13:438. [Crossref] [PubMed]
- Qiu J, Zhang T, Zhu X, et al. Hyperoside induces breast cancer cells apoptosis via ROS-mediated NF-κB signaling pathway. Int J Mol Sci 2019;21:131. [Crossref] [PubMed]


