
The expression, function, and utilization of Protamine1: a literature review
Introduction
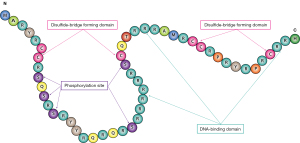
Protamine is a diverse family of small arginine-rich proteins that are synthesized in the late-stage spermatids of many animals and plants, including PRM1, PRM2, PRM3 in most species (1). PRMs bind to DNA leading to condensation of the spermatid chromatin and a genetically inactive status (1). Generally, the binding process is initiated by the genome-wide histone hyperacetylation and followed by the loading of spermatid-specific histone variant, H2A.L.2 onto the nucleosomes, and PRMs subsequently bind to DNAs to form an ultimately condensed sperm head through cooperating with transition proteins (TPs) (2,3). In certain species, a special type of protein also participates in the histone-protamine replacing process which is known as protamine-like proteins. The protamine-like proteins are the most heterogeneously sperm-basic nuclear proteins, possess higher amount of lysine and arginine residues than those of histones, and are larger than true protamine proteins (4,5). Structural analysis of the protamine-like proteins indicated that protamines are evolved from H1-like histones by a frameshift mutation in the carboxy-terminal end of the lysine-rich sperm-specific H1 histone (6-11). Compared with histones and protamine-like proteins, mature protamines are typically short proteins (50–100 amino acids) with higher amount of arginine (up to 70%) and two structural elements identified in all vertebrates. One element which contains multiple arginine or lysine amino acids, acts as small “anchoring” domains for DNA binding. The other element is comprised of multiple serine and threonine residues which are phosphorylation sites. Furthermore, in some species, a third domain exists with cysteine residues which can be oxidized to disulfide bridge to link them together and stabilize the chromatin complex (12).
Generally, primates and rodents express two types of protamines: PRM1 and PRM2. PRM1 is approximately 49–51 amino acids length and contains all the above-mentioned, highly evolutionary conserved domains (Figure 1). Whereas, protamine 2 (PRM2), with a species-specific expression manner, contains higher number of amino acids than PRM1 and presents in particular species. The human PRM1 and PRM2 genes co-locate in a tight cluster on chromosome 16 at 16p13.2 and the expressions of them are precisely regulated at transcription, translation, post-translation, and epigenetics levels (13,14). Noteworthily, transcription and translation of PRMs (PRM1 and PRM2) in mammalians, are temporarily uncoupled in the round spermatid stage and the late stage of spermatogenesis, respectively. This stage-specific pattern of gene expression is essential for the correct histone-protamine exchange and complete differentiation of round spermatids into mature spermatozoa.
So far, researches on male infertility have revealed that aberrant expression of PRM1 or ratio of PRM1/PRM2 created disrupted sperm shape or impaired sperm function which are responsible for male infertility (1,15), Besides, PRM1 is currently under investigation in various areas. For instance, the expression of PRM1 which belongs to the cancer-testis antigen (CTA) family (known as CT94.1), is upregulated in colorectal cancer (CRC) and chronic leukemia (CLL). Researches disclosed the cellular function of PRM1 in promoting CRC cell proliferation and metastasis, also detected the immunogenicity of PRM1 in CLL (16-18). Moreover, biofunction of PRM1 on cell growth is thought to be intricate. It has been reported that the increased rate of tumorigenesis was observed by subcutaneous injection of protamines (subtracted from salmon sperm), as well as the impaired proliferation of Hela cells and E. coli by repeated transfection of PcDNA-PRM1 (19-24). Additionally, due to the high affinity to DNA, PRM1 has been applied in various biomedicine fields such as transgenic animal research, development of drug delivery system, and gene therapy (20,25). Spermatid-like structures of somatic cell nucleus have been observed after heterologous expression of PRM1, which facilitates somatic cell nucleus transfer (SCNT) (19-21). Moreover, researchers applied peptide-derived approaches using PRM1 as a nucleic acid vector to protect and facilitate the delivery of siRNA or mRNA to specific cell populations (25-28). Within the scope of this review, we will illustrate the essential roles which PRM1 plays during mammalian spermatogenesis, and summarize the current research concerning PRM1 in other areas, aiming to broaden our present understandings of the biological significance of PRM1 and inspire further studies in tumor diagnosis, transgenic animal research, and novel drug delivery system in the future.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1582).
Biological function of PRM1
Pre-fertilization stage
Generally, during spermatogenesis, the mRNA of TPs and PRMs are stored in ribosomal protein granules of cytoplasm after transcription, followed by translation into proteins, and participate in sperm prolongation and spermatid differentiation (29,30). In mammalian spermatocytes, chromatin condensation is initiated by TPs and completed by PRMs (PRM1 and PRM2). During spermatogenesis, among the plenty mRNAs which expressed in round spermatids, the testis specific center and junction histone subtype expressed first, which possesses low affinity to DNA, leading to the nucleosome instability after post-translational modification. Subsequently, the acetylated histone is identified by bromodomain testis-specific proteins (Brdt), providing conditions for the coming TPs which work together with topoisomerase to relieve torsional stress. Translation of PRM1 is activated in elongating spermatids, simultaneously with the initiation of the final stage of chromatin condensation (29). Guided by TPs, individual PRM1 molecule binds to 10–11 bps of DNA, and PRM2 binds to a slightly larger segment of DNAs (around 15 bps) because the negative charge along the DNA backbone can be neutralized by PRM1 and PRM2, enabling adjacent DNA molecules to pack close together (3,31). Specifically, in the major groove, PRM1 wraps around the DNA helix, and one molecule being bound per turn of DNA helix. With the intra- and inter- molecular disulfide crosslinks formed by cysteine residues in PRM1, the characteristic toroidal structures are created and each contains roughly 50 kb of DNAs. This binding between PRM1, PRM2 and DNA backbone leads to the remarkable compression of approximately 85% of chromatins which are finally stored in the sperm head, and a small proportion of histones is retained which plays important roles after fertilization (32-34). These processes guarantee the unharmed transferring of the paternal genetic information to the oocyte and assure that the genetic information can be properly accessed by the developing embryo (1).
Post-fertilization stage
PRM1 is not only an essential protein in spermatogenesis, but the timely removal of PRM1 from DNA also affects the early development of embryo after fertilization. In human, it is reported that a programmed protamine-to-histone exchange occurs immediately after the paternal chromatin enters in the egg (35-37). Remarkably, the tightly packed chromatin from sperm is rapidly decondensed in assistance of proteins released from the oocyte nucleus. During this process, the chromatin from the parental side rapidly loss PRMs and testis specific histones, and is remodeled into the nucleosome structure with the help of histones from the mother. Recently, researchers have proposed and illustrated in detail the specific mechanism of protamine-to-histone exchange in the fertilized egg. In that report, the phosphorylation of PRM1 by SR (serine/arginine-rich domain) family of splicing factors protein kinase 1 (SRPK1) from the maternal side is essential for paternal chromatin decondensation and reprogramming (35). Noteworthily, the protamine-histone exchange has inspired more and more researches of PRM1 in other areas, such as transgenic animal research and drug delivery system development which will be illustrated later in this review.
Regulation mechanism of PRM1 expression
As early-described, PRMs participate in critical processes during spermatogenesis, and any abnormality of PRMs expression may affect the sperm parameter, and impair the shape or motility of sperm, finally leading to infertility (15,38,39). Therefore, the correct PRMs expression is believed to be a kind of chromatin checkpoint during sperm development. So far, accumulating studies have been carried out to survey the mechanism of PRM1 expression regulation in human which can be categorized into four levels.
Transcription level
So far, several mechanisms involved in the transcriptional regulation of PRM1 have been revealed, including association with nuclear matrix attachment regions (MARs), and binding of trans-acting factors to the promoter region (40,41). Firstly, MARs, located in the 5' and 3' regions around PRM1 genes, acts as cis-regulatory units in attachment of DNA to the protein scaffolding of the nuclear matrix. Research of MARs-knockout mouse models demonstrated that 3'-MAR can effectively promote gene activation (41). Pygopus family PHD finger 2 (PYGO2), belonging to a family of evolutionarily conserved PHD finger proteins, acts as co-activators of Wnt signaling. However, mice containing hypomorphic alleles of pygopus 2 revealed that mice are infertile with drastically spermatogenic defects and the expressions of Prm1, Prm2, Tnp2 and H1 fnt reduced remarkably (42). Transcription factor cAMP response element modulator (CREM), which is highly expressed in male germ cells, participates in regulation of many genes expression during spermatogenesis (43). Deletion of CREM gene in mice caused loss of TNPs and PRMs, and mice with camk4 gene deletion have been reported to display impaired spermatogenesis (44,45). Besides, TATA-box protein (TBP) and Y-box proteins are also important regulators of PRM1 expression during spermatogenesis. TBP over-expression is observed in mice between 18 and 28 days, corresponding to the period of the transcription of genes in haploid cells. Specifically, TBP-like factor (TLF), similar to TBP in sequence, may both activate and repress protamine transcription, and the TLF null mice were reported to have abnormal heterochromatin organization, which may affect histone-protamine exchange, chromatin condensation and even induces sperm apoptosis (46-48). Another important regulator of gene expression is Y-box protein, which binds both DNAs and RNAs to up-regulate transcription and down-regulate translation generally. So far, several Y-box proteins have been identified to regulate expression of PRM1. For instance, Contrin and its mouse orthologue have been reported to be associated with the PRMs expression. Contrin serves as a co-activator of PRMs transcription and can bind to the Y-box element in the gene promoter region. Additionally, Contrin polymorphism in population has been reported to be related to male infertility (49,50).
Translation level
Although PRM1 and PRM2 are transcribed in the round spermatid stage, their mRNAs are stored in translationally repressed ribonucleoproteins (RNPs) in the early haploid cells and activated in elongated spermatids (51,52). It is well-accepted that the expression of PRM1 and PRM2 are mainly under translational control, which is essential for correct sequential nucleoprotein exchange and complete differentiation of round spermatids into mature spermatozoa. Till now, the specific mechanism of PRM1 and PRM2 translational regulation is still yet to be fully illustrated. It is reported that the Y-box proteins Contrin and Translin which are mentioned above, also involve in PRM1 translational regulation (53). They are essential for RNP transportation to the cytoplasm and responsible for delayed translation (53-58). Specifically, Y-box protein 2 (YBX2) was identified to be responsible for repressed PRM1 mRNA translation because the depletion of YBX2 in Ybx-2 null mice aberrantly activates Prm1 translation prematurely in early spermatids (53). Additionally, transgenic animal research revealed that mice carrying a targeted disruption of Tarbp2 were sterile and severely oligospermic, proposing PRBP as a requisite for properly translational activation of the mRNAs encoding protamines (54,59,60).
Post-translation level
Different from PRM2 which is translated as a precursor demanding further splicing, PRM1 is translated as a mature protein without any cleavage, but rapid phosphorylation occurs soon after translation before binding to chromatin. As PRM1 and other PRMs are extended members of the SR family (61-63), they are regulated by the well-known evolutionarily conserved SRPKs (64,65). Among them, SRPK1, which is ubiquitously expressed in sperm cells, phosphorylates PRM1 at Ser10 and Ser8 soon after translation before entering the nucleus, which is essential for correct binding of PRM1 to DNA (35,66). Additionally, researchers also reported another significance of phosphorylation of SRPK1 after fertilization that it enhances and facilitates the removal of PRM1 from DNA after fertilization, which is a prerequisite for paternal chromatin decondensation in the fertilized egg and subsequent zygotic development (35).
Epigenetics level
JmjC-domain containing histone demethylase 2A (JHDM2A) is a H3K9 demethylase which is highly expressed in round spermatids. As reported by Okada et al., during mouse spermatogenesis, JHDM2A directly binds to the promoter regions of PRM1 and TNP1, and controls their expression by removing H3K9 at their promoters (67). Moreover, transgenic mice with Jhdm2a-deficient exhibit post-meiotic chromatin condensation defects and polymorphisms in exon 12 and exon 24 of JHDM2A gene are associated with male infertility (67-69).
Clinical utilization of PRM1
Being a famous molecular in spermatogenesis and fertilization, researches of PRM1 are accumulating. Abnormal expression of PRM1 is responsible for male infertility, and researchers also attempted to introduce PRM1 expression to somatic cells which inspired several studies concerning tumorigenesis, transgenic animal research, and drug delivery system (Figure 2).
Male infertility
Infertility is a global issue affecting approximately 15% of heterosexual couples, and male infertility factors accounts for 50% couples’ infertility (70). Generally, etiologies of male infertility can be categorized to four aspects: spermatogenic quantitative defects; ductal obstruction or dysfunction; hypothalamic-pituitary axis disturbances and spermatogenic qualitative defects (71). Since protamine proteins participate in the late stage of spermatogenesis, abnormality of PRM1 expression is responsible for spermatogenic qualitative defects. Accumulating studies have revealed the relationship between the abnormal mRNA levels or ratio of PRM1/PRM2 and sperm dysfunctions. It is reported recently that PRM1 and PRM2 mRNA copy numbers in the semen samples from teratozoospermic patients were much lower than normozoospermic patients and a normal PRM1/PRM2 mRNA (0.8–1.0) ratio was correlated with successful intracytoplasmic sperm injection cycles (72). Moreover, abnormal expression and ratio of PRM1/PRM2 were also observed in patients whose partners suffered the unexplained recurrent miscarriages (73). It is known that any single nucleotide polymorphism (SNP) in the coding or non-coding areas of PRM1 genes may lead to significant abnormalities in its expression, further impact the spermatogenesis processes and the products, which will ultimately lead to spermatogenesis dysfunction and male infertility (74). So far, researchers have screened PRM1 gene polymorphisms in infertility patients from different countries and districts (75-77). Collecting these studies, we find some variations related to male infertility, including the c.-109G>C, c.102G>T and c.119G>A in oligozoospermic men (78), c.-191C>A in oligozoospermic and asthenozoospermic men (79), and 230C>A in teratozoospermic infertile men (77). However, a recently published meta-analysis by Nemati et al. analyzed all the published articles about PRM1 and PRM2 polymorphisms, and evaluated the association with susceptibility to male infertility. By analysis, one previously reported type of PRM1 polymorphism as protective factors against Asian infertility was ruled out, and a subset of PRM1 polymorphisms was identified to be associated with elevated risk of male infertility (74).
Effect on tumorigenesis
The unique expression pattern in spermatid justifies PRM1 as a member of CTA family, which was first identified in CLL and CRC patients (16,18). PRM1 mRNA expression was detected in 11/41 (26.8%) early CLL patients, furthermore, high titers PRM1 IgG antibodies were also detectable in nearly half CLL patients but not in healthy donors (16). Additionally, analysis of CRC tissues also revealed increased expression level of PRM1 and explored the oncogenic function of PRM1 (18). However, the detailed biological functions of PRM1 in cancer are yet to be fully illustrated. Previous study has used subcutaneous injection of protamines exacted from salmon to detect the tumorigenesis rate in inbred rats which were given carcinogen dimethylhydrazine (DMH). The increased incidence of bowel tumor was observed in protamines treated group compared with heparin or 5-fluorouracil treated groups (23). Furthermore, researchers are inspired by the special histone-protamine exchange process which leads to chromatin condensation and transcriptional termination in spermatids. For instance, Günther et al. analyzed the protamine-mediated chromatin condensation in somatic cancer cell line (22). Through repeat transfection of PcDNA-PRM1 or PcDNA-PRM2, they observed the impaired proliferation in two fast-growing cell systems, E. coli and Hela cells, and detected the foci of PRM1 and PRM2 distributed within the nucleus of Hela cells. Collectively, as a CTA, PRM1 possess a highly tissue-specific expression pattern, and induce certain immune response. The above-mentioned results may inspire future researches to investigate PRM1 expression and functions as a novel CTA in other human malignancies.
Application in transgenic animal research
Although SCNT is a promising technology in transgenic animal research, the full implementation is still on hold because of the low efficiency (80,81). The strategies targeting nuclear reprogramming include the RNAi-mediated downregulation of Xist, the depletion of H3K9 and H3K27 methyltransferases in somatic cells, and modulation of DNA methylation by dCas9-Tet/Dnmt before nucleus transfer (NT) (82-85). Recently, Iuso et al. have developed a novel protocol aiming to facilitate the SCNT procedures. The protocol is to introduce PRM1 expression into adult somatic cells through transfection methods, overexpression of PRM1 was observed in somatic cells. When the overexpressed PRM1 protein entered the nuclei, histone-protamine exchange occurred in the chromatin which was formed into condensed station, a spermatid-like structures. Subsequently, researchers introduced the spermatid-like somatic cell nuclei into enucleated oocytes, and observed the reversible remodeling process which resulted into normally developed embryos in vitro. Now this protocol is believed to be simpler, easily readable by the oocytes, and ultimately improve SCNT efficiency (19-21).
Novel vector for drug delivery system
Due to the high affinity to DNA backbone, the positive-charged PRMs, especially PRM1, is found to be great help in facilitating the development of novel gene carriers to selected cell populations (25-28,86-88). Lately, Weide et al. developed a more feasible and safe method to deliver tumor vaccine. The protamine-mRNA vaccine coding tumor-associated antigens was constructed to induce antitumor immune responses, and has achieved satisfactory effects on melanoma patients (26). Meanwhile, utilization of the small interfering RNAs (siRNAs)-targeted certain oncogenic genes or proteins might turn out a promising anticancer therapy to retard cancer cell growth and metastasis. However, the efficient delivery of siRNA into cancer cells in vivo remains frustrating, and many strategies have been applied to improve the efficacy in several malignancies (89-91). Researchers took advantage of the reversible binding process of PRMs to nucleic backbone, and developed fusion proteins of single-chain fragmented antibodies and protamine-derived peptides, which is used to deliver siRNAs into specific target cells and proved to be more effective methods than conventional delivery (28). Moreover, protamine-derived peptide has been widely used to develop efficient biodegradable non-viral carrier for gene transfection, in conjugation with other materials, which has promising prospect in nanotechnology (86,92,93).
Conclusions
Spermatogenesis and fertilization are the primary assurance of life development. Sperm at fertilization transfers not only its genetic but also epigenetic information. PRM1 and PRM2 play important roles in these processes, which have been proposed as a suitable biomarker for estimating male sperm quality, especially for male infertility. Therefore, although PRM1 is a well-known protein in andrology research, further studies are extremely expected which will reveal the underlying mechanism of PRM1 in chromatin remodeling and explore the clinical significance in male infertility.
On the other hand, PRM1 belongs to CTA family, which is aberrantly expressed in some malignancies and induces specific humoral immune response. It will be rather promising to develop PRM1-based tumor diagnosis and treatment strategies. Besides, the high-titers of PRM1-specific IgG in CLL patients also expands our horizon of its employment in more translational clinical researches as well as the nucleic acid delivery vector or transgenic animal research. Till now articles are limited concerning the expression and biological function of PRM1 in cancer or other diseases, partly due to the facts of heterogenous expression pattern of CTA and the special remodeling change of chromatin after exogenous overexpression of PRM1 which may poses obstacles for conventional research of PRM1 in tumors. In conclusion, PRM1 represents not only an essential checkpoint for sperm chromatin quality control but also a novel target for tumor diagnosis and biotherapy. Future studies are long expected to clarify the underlying mechanism in spermatogenesis and illustrate detailed cellular functions of PRM1 in tumorigenesis and metastasis.
Acknowledgments
The authors would like to thank Director of National Natural Science Foundation of China for providing funding for the research (grant No. 31771093). The authors would also like to thank Dr. Yan Ding for writing assistance.
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1582
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-1582
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1582). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Steger K, Balhorn R. Sperm nuclear protamines: A checkpoint to control sperm chromatin quality. Anat Histol Embryol 2018;47:273-9. [Crossref] [PubMed]
- Goudarzi A, Shiota H, Rousseaux S, et al. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J Mol Biol 2014;426:3342-9. [Crossref] [PubMed]
- Barral S, Morozumi Y, Tanaka H, et al. Histone Variant H2A.L.2 Guides Transition Protein-Dependent Protamine Assembly in Male Germ Cells. Mol Cell 2017;66:89-101.e8. [Crossref] [PubMed]
- Lewis JD, Ausió J. Protamine-like proteins: evidence for a novel chromatin structure. Biochem Cell Biol 2002;80:353-61. [Crossref] [PubMed]
- D'Ippolito RA, Minamino N, Rivera-Casas C, et al. Protamines from liverwort are produced by post-translational cleavage and C-terminal di-aminopropanelation of several male germ-specific H1 histones. J Biol Chem 2019;294:16364-73. [Crossref] [PubMed]
- Lewis JD, Saperas N, Song Y, et al. Histone H1 and the origin of protamines. Proc Natl Acad Sci U S A 2004;101:4148-52. [Crossref] [PubMed]
- Kasimatis KR, Moerdyk-Schauwecker MJ, Timmermeyer N, et al. Proteomic and evolutionary analyses of sperm activation identify uncharacterized genes in Caenorhabditis nematodes. BMC Genomics 2018;19:593. [Crossref] [PubMed]
- de Vries JC, Barendrecht AD, Clark CC, et al. Heparin Forms Polymers with Cell-free DNA Which Elongate Under Shear in Flowing Blood. Sci Rep 2019;9:18316. [Crossref] [PubMed]
- Eirín-López JM, Frehlick LJ, Ausió J. Protamines, in the footsteps of linker histone evolution. J Biol Chem 2006;281:1-4. [Crossref] [PubMed]
- Eirín-López JM, Lewis JD. Common phylogenetic origin of protamine-like (PL) proteins and histone H1: Evidence from bivalve PL genes. Mol Biol Evol 2006;23:1304-17. [Crossref] [PubMed]
- Martínez-Soler F, Kurtz K, Ausió J, et al. Transition of nuclear proteins and chromatin structure in spermiogenesis of Sepia officinalis. Mol Reprod Dev 2007;74:360-70. [Crossref] [PubMed]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol 2007;8:227. [Crossref] [PubMed]
- Schneider S, Balbach M, Jan F. Re-visiting the Protamine-2 locus: deletion, but not haploinsufficiency, renders male mice infertile. Sci Rep 2016;6:36764. [Crossref] [PubMed]
- Wykes SM, Krawetz SA. Conservation of the PRM1 --> PRM2 --> TNP2 domain. DNA Seq 2003;14:359-67. [Crossref] [PubMed]
- Pardede BP, Agil M, Supriatna I. Protamine and other proteins in sperm and seminal plasma as molecular markers of bull fertility. Vet World 2020;13:556-62. [Crossref] [PubMed]
- Meklat F, Zhang Y, Shahriar M, et al. Identification of protamine 1 as a novel cancer-testis antigen in early chronic lymphocytic leukaemia. Br J Haematol 2009;144:660-6. [Crossref] [PubMed]
- Lim SH, Zhang Y, Zhang J. Cancer-testis antigens: the current status on antigen regulation and potential clinical use. Am J Blood Res 2012;2:29-35. [PubMed]
- Chen Z, Shi C, Gao S, et al. Impact of protamine I on colon cancer proliferation, invasion, migration, diagnosis and prognosis. Biol Chem 2018;399:265-75. [Crossref] [PubMed]
- Iuso D, Czernik M, Toschi P, et al. Exogenous Expression of Human Protamine 1 (hPrm1) Remodels Fibroblast Nuclei into Spermatid-like Structures. Cell Rep 2015;13:1765-71. [Crossref] [PubMed]
- Palazzese L, Czernik M, Iuso D, et al. Nuclear quiescence and histone hyper-acetylation jointly improve protamine-mediated nuclear remodeling in sheep fibroblasts. PLoS One 2018;13:e0193954. [Crossref] [PubMed]
- Czernik M, Iuso D, Toschi P, et al. Remodeling somatic nuclei via exogenous expression of protamine 1 to create spermatid-like structures for somatic nuclear transfer. Nat Protoc 2016;11:2170-88. [Crossref] [PubMed]
- Günther K, Paradowska-Dogan A, Bärmann B, et al. Expression of sperm-specific protamines impairs bacterial and eukaryotic cell proliferation. Histochem Cell Biol 2015;143:599-609. [Crossref] [PubMed]
- Phillips RK, Quill DS, Dudley HA. Experimental colon neoplasia enhanced by extract of sperm (protamine). J R Soc Med 1984;77:830-2. [Crossref] [PubMed]
- Klabukova DL, Labutina NV, Vasilievich NV, et al. Protamine Impact on Viability of Bacterial and Eukaryotic Cell Cultures. Asian J Pharm 2017;11:S504-9.
- Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 2020;19:673-94. [Crossref] [PubMed]
- Weide B, Pascolo S, Scheel B, et al. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother 2009;32:498-507. [Crossref] [PubMed]
- Pardi N, Hogan MJ, Porter FW, et al. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov 2018;17:261-79. [Crossref] [PubMed]
- Yao YD, Sun TM, Huang SY, et al. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci Transl Med 2012;4:130ra48. [Crossref] [PubMed]
- Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update 2007;13:313-27. [Crossref] [PubMed]
- Bashiri Z, Amidi F, Amiri I, et al. Male Factors: the Role of Sperm in Preimplantation Embryo Quality. Reprod Sci 2021;28:1788-811. [Crossref] [PubMed]
- Staub C, Johnson L. Review: Spermatogenesis in the bull. Animal 2018;12:s27-35. [Crossref] [PubMed]
- Kobayashi Y, Tomizawa SI, Ono M, et al. Tsga8 is required for spermatid morphogenesis and male fertility in mice. Development 2021;148:dev196212. [Crossref] [PubMed]
- Hecht N, Behr R, Hild A, et al. The common marmoset (Callithrix jacchus) as a model for histone and protamine expression during human spermatogenesis. Hum Reprod 2009;24:536-45. [Crossref] [PubMed]
- Chioccarelli T, Pierantoni R, Manfrevola F, et al. Histone Post-Translational Modifications and CircRNAs in Mouse and Human Spermatozoa: Potential Epigenetic Marks to Assess Human Sperm Quality. J Clin Med 2020;9:640. [Crossref] [PubMed]
- Gou LT, Lim DH, Ma W, et al. Initiation of Parental Genome Reprogramming in Fertilized Oocyte by Splicing Kinase SRPK1-Catalyzed Protamine Phosphorylation. Cell 2020;180:1212-1227.e14. [Crossref] [PubMed]
- Adenot PG, Szöllösi MS, Geze M, et al. Dynamics of paternal chromatin changes in live one-cell mouse embryo after natural fertilization. Mol Reprod Dev 1991;28:23-34. [Crossref] [PubMed]
- Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 2014;30:581-613. [Crossref] [PubMed]
- Oliva R. Protamines and male infertility. Hum Reprod Update 2006;12:417-35. [Crossref] [PubMed]
- Jodar M, Oliva R. Protamine alterations in human spermatozoa. Adv Exp Med Biol 2014;791:83-102. [Crossref] [PubMed]
- Ward WS. Organization of sperm DNA by the nuclear matrix. Am J Clin Exp Urol 2018;6:87-92. [PubMed]
- Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J Biol Chem 2004;279:51862-8. [Crossref] [PubMed]
- Nair M, Nagamori I, Sun P, et al. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev Biol 2008;320:446-55. [Crossref] [PubMed]
- Wang C, Chen Y, Manthari RK, et al. Abnormal spermatogenesis following sodium fluoride exposure is associated with the downregulation of CREM and ACT in the mouse testis. Toxicol Ind Health 2018;34:219-27. [Crossref] [PubMed]
- Wu JY, Ribar TJ, Cummings DE, et al. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet 2000;25:448-52. [Crossref] [PubMed]
- Kosir R, Juvan P, Perse M, et al. Novel insights into the downstream pathways and targets controlled by transcription factors CREM in the testis. PLoS One 2012;7:e31798. [Crossref] [PubMed]
- Yu C, Cvetesic N, Hisler V, et al. TBPL2/TFIIA complex establishes the maternal transcriptome through oocyte-specific promoter usage. Nat Commun 2020;11:6439. [Crossref] [PubMed]
- Martianov I, Brancorsini S, Gansmuller A, et al. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development 2002;129:945-55. [Crossref] [PubMed]
- Martianov I, Fimia GM, Dierich A, et al. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 2001;7:509-15. [Crossref] [PubMed]
- Aydos OSE, Hekmatshoar Y, Altınok B, et al. Genetic Polymorphisms in PRM1, PRM2, and YBX2 Genes are Associated with Male Factor Infertility. Genet Test Mol Biomarkers 2018;22:55-61. [Crossref] [PubMed]
- Yang J, Medvedev S, Yu J, et al. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci U S A 2005;102:5755-60. [Crossref] [PubMed]
- Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res 2003;103:217-24. [Crossref] [PubMed]
- Kleene KC. Connecting cis-elements and trans-factors with mechanisms of developmental regulation of mRNA translation in meiotic and haploid mammalian spermatogenic cells. Reproduction 2013;146:R1-19. [Crossref] [PubMed]
- Cullinane DL, Chowdhury TA, Kleene KC. Mechanisms of translational repression of the Smcp mRNA in round spermatids. Reproduction 2015;149:43-54. [Crossref] [PubMed]
- Oyama K, Baba T, Kashiwabara SI. Functional characterization of testis-brain RNA-binding protein, TB-RBP/Translin, in translational regulation. J Reprod Dev 2021;67:35-42. [Crossref] [PubMed]
- Gupta A, Pillai VS, Chittela RK. Translin: A multifunctional protein involved in nucleic acid metabolism. J Biosci 2019; [Crossref] [PubMed]
- Kwon YK, Hecht NB. Cytoplasmic protein binding to highly conserved sequences in the 3' untranslated region of mouse protamine 2 mRNA, a translationally regulated transcript of male germ cells. Proc Natl Acad Sci U S A 1991;88:3584-8. [Crossref] [PubMed]
- Mordovkina D, Lyabin DN, Smolin EA, et al. Y-Box Binding Proteins in mRNP Assembly, Translation, and Stability Control. Biomolecules 2020;10:591. [Crossref] [PubMed]
- Murray MT, Schiller DL, Franke WW. Sequence analysis of cytoplasmic mRNA-binding proteins of Xenopus oocytes identifies a family of RNA-binding proteins. Proc Natl Acad Sci U S A 1992;89:11-5. [Crossref] [PubMed]
- Zhong J, Peters AH, Lee K, et al. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet 1999;22:171-4. [Crossref] [PubMed]
- Chennathukuzhi V, Morales CR, El-Alfy M, et al. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci U S A 2003;100:15566-71. [Crossref] [PubMed]
- Zhong XY, Wang P, Han J, et al. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 2009;35:1-10. [Crossref] [PubMed]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol 2007;623:107-22. [Crossref] [PubMed]
- Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 2015;6:93-110. [Crossref] [PubMed]
- Giannakouros T, Nikolakaki E, Mylonis I, et al. Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J 2011;278:570-86. [Crossref] [PubMed]
- Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013;122:191-207. [Crossref] [PubMed]
- Papoutsopoulou S, Nikolakaki E, Chalepakis G, et al. SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res 1999;27:2972-80. [Crossref] [PubMed]
- Okada Y, Scott G, Ray MK, et al. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007;450:119-23. [Crossref] [PubMed]
- Hojati Z, Nouri Emamzadeh F, Dehghanian F. Association between polymorphisms of exon 12 and exon 24 of JHDM2A gene and male infertility. Int J Reprod Biomed 2016;14:389-96. [Crossref] [PubMed]
- Okada Y, Tateishi K, Zhang Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J Androl 2010;31:75-8. [Crossref] [PubMed]
- Krausz C, Escamilla AR, Chianese C. Genetics of male infertility: from research to clinic. Reproduction 2015;150:R159-74. [Crossref] [PubMed]
- Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol 2018;15:369-84. [Crossref] [PubMed]
- Savadi-Shiraz E, Edalatkhah H, Talebi S, et al. Quantification of sperm specific mRNA transcripts (PRM1, PRM2, and TNP2) in teratozoospermia and normozoospermia: New correlations between mRNA content and morphology of sperm. Mol Reprod Dev 2015;82:26-35. [Crossref] [PubMed]
- Rogenhofer N, Ott J, Pilatz A, et al. Unexplained recurrent miscarriages are associated with an aberrant sperm protamine mRNA content. Hum Reprod 2017;32:1574-82. [Crossref] [PubMed]
- Nemati H, Sadeghi M, Nazeri M, et al. Evaluation of the association between polymorphisms of PRM1 and PRM2 and the risk of male infertility: a systematic review, meta-analysis, and meta-regression. Sci Rep 2020;10:17228. [Crossref] [PubMed]
- Jiang WJ, Zhang J, Xia XY, et al. Protamine gene polymorphisms and male infertility. Zhonghua Nan Ke Xue 2015;21:1134-7. [PubMed]
- Grassetti D, Paoli D, Gallo M, et al. Protamine-1 and -2 polymorphisms and gene expression in male infertility: an Italian study. J Endocrinol Invest 2012;35:882-8. [PubMed]
- Dehghanpour F, Fesahat F, Miresmaeili SM, et al. Analysis of PRM1 and PRM2 Polymorphisms in Iranian Infertile Men with Idiopathic Teratozoospermia. Int J Fertil Steril 2019;13:77-82. [PubMed]
- Ravel C, Chantot-Bastaraud S, El Houate B, et al. Mutations in the protamine 1 gene associated with male infertility. Mol Hum Reprod 2007;13:461-4. [Crossref] [PubMed]
- Abdallah WI, Hussein TM, Elsayed ET, et al. The c.-190 C>A transversion in promoter region of protamine 1 gene as a genetic risk factor in Egyptian men with idiopathic infertility. Andrologia 2019;51:e13367. [Crossref] [PubMed]
- Czernik M, Anzalone DA, Palazzese L, et al. Somatic cell nuclear transfer: failures, successes and the challenges ahead. Int J Dev Biol 2019;63:123-30. [Crossref] [PubMed]
- Wilmut I, Schnieke AE, McWhir J, et al. Viable offspring derived from fetal and adult mammalian cells. Nature 1997;385:810-3. [Crossref] [PubMed]
- Matoba S, Zhang Y. Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell Stem Cell 2018;23:471-85. [Crossref] [PubMed]
- Matoba S, Inoue K, Kohda T, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci U S A 2011;108:20621-6. [Crossref] [PubMed]
- Matoba S, Liu Y, Lu F, et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 2014;159:884-95. [Crossref] [PubMed]
- Zhou C, Wang Y, Zhang J, et al. H3K27me3 is an epigenetic barrier while KDM6A overexpression improves nuclear reprogramming efficiency. FASEB J 2019;33:4638-52. [Crossref] [PubMed]
- Priya SS, Rekha MR, Sharma CP. Pullulan-protamine as efficient haemocompatible gene delivery vector: synthesis and in vitro characterization. Carbohydr Polym 2014;102:207-15. [Crossref] [PubMed]
- Delgado D, Gascón AR, Del Pozo-Rodríguez A, et al. Dextran-protamine-solid lipid nanoparticles as a non-viral vector for gene therapy: in vitro characterization and in vivo transfection after intravenous administration to mice. Int J Pharm 2012;425:35-43. [Crossref] [PubMed]
- Holder E, Griesenbach U, Li S, et al. Intravenously administered oligonucleotides can be delivered to conducting airway epithelium via the bronchial circulation. Gene Ther 2006;13:1628-38. [Crossref] [PubMed]
- Zhang J, Du Z, Pan S, et al. Overcoming Multidrug Resistance by Codelivery of MDR1-Targeting siRNA and Doxorubicin Using EphA10-Mediated pH-Sensitive Lipoplexes: In Vitro and In Vivo Evaluation. ACS Appl Mater Interfaces 2018;10:21590-600. [Crossref] [PubMed]
- Liu HY, Yu X, Liu H, et al. Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Sci Rep 2016;6:30346. [Crossref] [PubMed]
- Bäumer S, Bäumer N, Appel N, et al. Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clin Cancer Res 2015;21:1383-94. [Crossref] [PubMed]
- Arabzadeh S, Amiri Tehranizadeh Z, Moalemzadeh Haghighi H, et al. Design, Synthesis, and In Vitro Evaluation of Low Molecular Weight Protamine (LMWP)-Based Amphiphilic Conjugates as Gene Delivery Carriers. AAPS PharmSciTech 2019;20:111. [Crossref] [PubMed]
- Ji X, Wang C, Tang M, et al. Biocompatible protamine sulfate@silicon nanoparticle-based gene nanocarriers featuring strong and stable fluorescence. Nanoscale 2018;10:14455-63. [Crossref] [PubMed]



