
Advances in the phase separation-organized membraneless organelles in cells: a narrative review
Introduction
Compartmentalization of cells into distinct, functional rooms in a space- and time-regulated manner is essential in eukaryotic cells, facilitating spatiotemporal regulation of biological reactions. In these separate compartments, there are a multitude of complex biochemical reactions over diverse, active components to carry out various physiological processes (1,2). With the influence of the surrounding environment, cells rapidly regulate the rate and direction of these reactions based on the localization of the reaction components. Specifically, concentrating components within a specific space can significantly increase the reaction kinetics to meet physiological needs, whereas segregating them can remarkably slow or inhibit reactions to protect cells from damaging activities under adverse environmental conditions, such as stress, hypoxia, and low pH (3-5).
Membrane-bound organelles (MOs) are subcellular compartments separated from their surrounding environment by an apparent boundary, orchestrated by surrounding lipid bilayer membranes, and they include the nucleus, mitochondria, lysosomes, endoplasmic reticulum, and Golgi apparatus (6). In addition to canonical MOs, cells also harbor several novel compartments that have no obvious physical boundaries, which is attributed to the lack of a delimiting membrane. Interestingly, membraneless organelles (MLOs) contain a high concentration of internal components, including proteins, nucleic acids, and other molecules (7-9). Recently, MLOs have attracted considerable interest from researchers due to their potential roles in multiple biological processes. Based on the phase-separation phenomenon, MLOs may endow cells with the ability of self-protection in response to environmental change, including the regulation of gene expression, the stability of protein, and the control of signal transduction (10-14). In this review, we comprehensively elucidated the formation of these MLOs and the relationship between biological functions and associated diseases. The mechanisms underlying the influence of protein concentration and valency on phase separation in cells are also discussed.
MLOs and biomolecular phase separation
In the field of cell biology, MLOs are more generally known as biomolecular condensates, in which the inner molecules are concentrated to a higher concentration relative to their surrounding milieu (15). Interestingly, recent studies have highlighted that the driving force underlying the formation of MLOs is mainly generated by the liquid-liquid phase separation (LLPS) microenvironment (16). Phase transition is a well-known process that spontaneously occurs when the concentration of the components reaches a certain threshold, and is ubiquitous and serves a vital function within living cells (Figure 1A). In particular, LLPS within the cells is thought to be the main driving force behind the formation of biomolecular condensates, in which several proteins, RNAs, and other biomolecules undergo phase separation and finally form liquid-like droplets, thereby becoming an important organizing principle for MLOs and further determining the organism responses following the changes in the external environment (17).
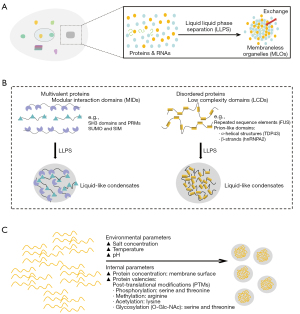
Protein structure and LLPS
Generally, internal molecules have several attractive structures that promote the interactions between components to drive the formation of condensed biomolecules (18). There are two main types of structural proteins involved in various biomolecular aggregates, including various proteins carrying different types of multiple modular interaction domains (MIDs) and proteins containing a large number of low complexity domains (LCDs) (Figure 1B) (19,20).
There are now several examples of proteins with structural characteristic properties of multiple MIDs that drive phase separation, which is proved to be important for signaling complexes. For example, the actin-regulatory signaling pathway has been described, and mainly contain three types of multivalent proteins: phospho-tyrosine residues of nephrin, Src homology 2 (SH2), and SH3 domains of Nck, and proline-rich motifs (PRMs) of neural Wiskott-Aldrich syndrome protein (N-WASP) (20). When the signaling pathway is activated, these proteins rapidly form phase-separated clusters through multivalent interactions of the internal components, such as between phospho-tyrosine residues and the SH2 domains and between PRMs and the SH3 domains. A recent study by Banani demonstrated that in various condensates, such as promyelocytic leukemia (PML) bodies, proteins comprising multiple repeats of small ubiquitin-related modifier (SUMO) domains could bind to other proteins with SUMO-interacting motif (SIM) ligands, thereby generating liquid-like condensates (21,22).
A second possible structure for condensate formation by proteins contains large LCDs, also named intrinsically disordered regions (IDRs) in most intracellular condensates. The driving force for phase separation of IDR-containing proteins can be provided by the multivalent interactions between proteins carrying with oppositely charged residues or between two molecules harboring repeated sequence elements. Kato showed that phase separation of RNA-binding proteins (RBPs) fused in sarcoma (FUS) was dependent on tyrosine residues enriched in FUS LCDs, and Pak demonstrated that many complementary charged residues were required for phase separation in the disordered intracellular domain of nephrin (23,24). In addition, proteins containing prion-like domains are the second category in aggregated proteins with many LCDs, including α-helical structures (such as TDP43 LCDs) and β-strands (such as hnRNPA2 LCDs), indicating the vital functions of secondary structures of proteins in the formation of liquid-like condensates (25). Conicella et al. elucidated that the driving force derived from weak multivalent adhesions of α-helical structures is important in the phase separation of TDP43 (26). Similarly, a recent study indicated that the interactions between β-strands could trigger the aggregation of hnRNPA2 proteins undergoing the LLPS process (27). In summary, protein-protein interactions based on repetitive sequences (MIDs or LCDs) are an essential driving force for the formation of MLOs by the LLPS microenvironment.
Regulation of phase separation in cellular compartmentalization
LLPS is an essential process underlying the formation of MLOs by multivalent interactions among proteins and nucleic acids. Accumulating evidence has revealed that there are several different types of multivalent interactions facilitating phase separation, regulated by various environmental parameters (such as salt concentration, temperature, and pH) and phase separation threshold of internal molecules (such as protein concentrations and valencies) (Figure 1C) (15,28). Next, we review the mechanisms underlying the influence of protein concentration and valency on phase separation in cells.
Control of cellular concentration
Generally, membrane surface comprising a lipid bilayer can effectively enable regulation over the local concentrations via surface tension compared to the cytosol as well as restrict the diffusion of biomolecules to two dimensions, thereby altering threshold concentrations at which the aggregates are easy to form through phase separation (29). Accumulation of proteins at the membrane surfaces has been confirmed in several signaling cascades, thus facilitating signal transduction. For instance, the transmembrane adaptor protein of T cells, termed linker for the activation of T cells (LAT), is essential for the activation of signaling downstream of the T cell receptor (TCR) (30). Recent studies found that following TCR activation upon engagement of the associated ligands, this protein network showed a trend for triggering phase separates, which was dependent on the formation of membrane-bound clusters, including Grb2 and SOS proteins, involved in intracellular domains of TCR (31).
Control of multivalent interactions
In addition to the concentration, post-translational modifications (PTMs) of proteins, including phosphorylation of serine and threonine, methylation of arginine, acetylation of lysine, and glycosylation of serine and threonine modified with O-linked N-acetylglucosaminylation (O-Glc-NAc), are largely implicated in the modulation of the phase separation process (32). Specifically, phosphorylation of proteins can enhance or suppress phase separation, underlining the specific regulatory effects of PTMs on different condensates, including FUS, CPEB4, and TDP43 proteins (33-35). Arginine methylation has been shown to suppress phase separation via impairing cation-p interactions in several RBPs, including FUS, RNA helicase Ddx4, and hnRNPA2 (36,37). Lysine acetylation has been shown to have differing effects on phase separation in different cases, such as inhibiting phase separation in Tau protein and promoting acetylation of TDP43 (38,39). Glycosylation (O-Glc-NAc) can impair phase separation of associated proteins, decreasing the formation of protein aggregates, such as Tau, hnRNPA1, and α-synuclein (40-42).
In summary, phase separation is an intrinsic feature of several biomolecules at sufficient concentration, including proteins and nucleic acids with various valencies. The cellular mechanisms that regulate phase separation include enrichment of internal molecules and regulation of PTMs, including phosphorylation, methylation, acetylation, and glycosylation. Hence, cells can control the phase separation threshold of internal molecules through regulating membrane surface tension and PTM of proteins, thereby forming several functional condensates at the correct time and location in pathological processes. We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1111).
Several membraneless-organelles and associated functions in cells
MLOs produced by LLPS is mainly distributed in the nucleus, nuclear membrane, cytoplasm, and plasma membrane. Here, we comprehensively discuss the physical principles and functional properties of each phase-separated MLO (Figure 2). A review of literature was conducted in PubMed, to identify the latest research on membraneless-organelles and associated functions in cells, and ultimately to generate a narrative review.
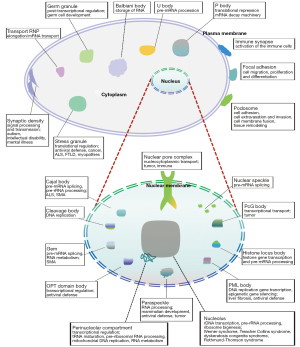
Nucleus
Cajal body (CB)
CBs, originally termed nucleolar accessory bodies or coiled bodies, are intranuclear MLOs known as a nuclear center for the assembly of ribonucleoproteins (RNPs), including small nuclear RNPs (snRNPs) and small nucleolar RNPs (snoRNPs) (61,62). Coilin is an essential scaffold protein of CBs, determining the structural integrity and function of CBs. The survival motor neuron (SMN) protein complex can form snRNPs in the cytoplasm and accompany them into the nucleus, and then has a tendency to release from the snRNPs for cycling back into the cytoplasm (63). A positive correlation has been reported between transcription rates and CB numbers in HeLa cells (64). Fluorescence recovery after photobleaching (FRAP) assays showed that several CB proteins had a good characteristic of rapid exchange with the nucleoplasm, especially coilin and SMN (65). Therefore, CBs possess dynamic behaviors in various cells, resulting in the variable activity of CBs in response to gene transcription-associated changes.
CBs promote the formation of ‘transcriptomes’, which is attributed to the assembly of several transcript factors associated with transcription, capping, splicing, polyadenylation, and cleavage of pre-mRNAs. Pellizzoni suggested that the SMN complex facilitated the accumulation of RNA pol II in coilin-containing structures due to the interaction of the C-terminal domain of RNA pol II and Gemins through RNA helicase A (66). According to previous studies, CB dysfunction is associated with multiple neuropathological disorders, such as the mutation of the SMN1 gene in amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy (SMA) (43).
Cleavage body
Cleavage bodies are nuclear sub-organelles that contain cleavage stimulation factor CstF-64 and polyadenylation specificity factor CPSF-100 (67). Previous studies showed that cleavage bodies were spatially distributed in various cell cycles, CstF-64-containing cleavage bodies were mainly enriched in the S phase while CPSF-100-containing cleavage bodies were enriched in S and G2 phases. Li suggested that inhibition of DNA replication in cells treated with hydroxyurea could eliminate the majority of CstF-64-containing cleavage bodies, further verifying the function of cleavage bodies in DNA replication rather than RNA transcription (68). The number of cleavage bodies increased while the level of CstF-64 proteins showed no significant change during DNA replication, indicating that the formation of CstF-64-containing cleavage bodies facilitated the redistribution of CstF-64 proteins within the nucleus.
Gem
Gems or Gemini of CBs are liquid-state aggregates of SMN complexes secreted from CBs in the nucleus. Gems are ‘storage cabin’ for excess nuclear SMN complexes ready for recycling into the cytoplasm and facilitate the formation of snRNPs following their participation in pre-mRNA splicing. Although gems usually overlap with CBs, CB contains both coilin and SMN whereas gem only comprises of SMN. Several studies showed that a high concentration of SMN complexes facilitated the formation of gem organelles; however, gem numbers tended to decrease significantly following the low level of SMN complexes (69). It is well known that SMN is the determining factor for SMA, a genetic condition associated with neuromuscular degenerative disorder (44).
Nuclear speckle (NS)
NSs are one of the most prominent nuclear MLOs containing many pre-mRNA splicing factors [such as snRNPs and serine/arginine-rich (SR) proteins] and are widely distributed in the interchromatin regions of the nucleoplasm (70). Although the process of transcription is not associated with NSs, there are various transcription factors, such as serine-2-phosphorylated RNA pol II, and associated transcription elongation proteins other than splicing factors within NSs, revealing the potential function of NS in transcriptional regulation and processing (71). The phosphorylation-dephosphorylation procedure is essential for pre-mRNA splicing through mediating the formation of the spliceosome complex (72). The reduction of phosphorylated SR proteins involved in NSs inhibits the spliceosome assembly; however, the increase in the level of phosphorylation relieves this block in the spliceosome assembly. The size and number of NSs increased due to the increased accumulation of splicing factors when transcription is halted by inhibitors or heat shock (73). In addition, splicing factors tended to leave NSs when gene transcription was active (74). In conclusion, NSs are essential organelles containing high concentrations of splicing snRNPs and other splicing-related factors and facilitate gene transcription and RNA processing within the nucleus.
Nucleolus
The nucleolus, a distinct subnuclear MLO assembled around chromosomal nucleolus organizer regions (NOR) composed of the repeated ribosomal DNA (rDNA) clusters, is a suitable compartment for rDNA transcription, pre-rRNA processing, and initial assembly and maturation of pre-ribosome (45,75,76). Recent studies have uncovered other essential functions of the nucleolus, including the regulation of multiple RNP assembly and PTMs of nuclear proteins, such as sumoylation and phosphorylation (77). The nucleolus also plays a special role in regulating multiple aspects of cell cycle progression through inhibiting activities of various specific proteins by sequestering these proteins in a specific compartment. Nucleolus undergoes sequential alternating cycles of disassembly and reorganization during mammalian cell mitosis, which is consistent with the view that the nucleolus is a dynamic structure; the disassembly of the nucleolus compartment turns off when mammalian cells enter mitosis and gene transcription (78). Given that the nucleolus is a ribosome production factory maintaining protein-synthesis levels during cell growth and division, the activity of the nucleolus was tightly regulated by various signaling events in cell-cycle regulation and stress responses (79). The nucleolus exerts its biological functions through regulating the level of tumor-suppressor protein p53, which coordinates cellular response to stress (80). Increasing evidence has also revealed that nucleolar structure or ribosome biogenesis is likely to change to inhibit cancer progression and viral infection (81).
OPT domain body
There is a special subnuclear MLO, termed Oct1/PTF/transcription (OPT) domains, that appear during the G1 stage of the cell division but disappear during the S stage, and contain several transcription factors such as PTF, Oct1, TBF, and Sp1 (82,83). PTF and Oct1 are regarded as useful switches to facilitate the expression of snRNAs and other ‘processing RNAs’ together with RNA pol II or III. Sp1 is a promoter-specific transcription factor that plays an essential role during cell metastasis and cell-cycle regulation by binding to GC-rich decanucleotide recognition elements. Interestingly, based on various transcription factors involved in OPT domain bodies, there are several important characteristics, including a high dynamic compartment at the transcription sites due to some highly disordered protein of transcription factors (84).
PcG body
Generally, positioning target genes into a special nuclear compartment, termed Polycomb group (PcG) body, induced gene repression due to repressive transcription-complexes, PcG repressive complexes 1 and 2 (PRC1 and PRC2), containing numerous gene-silencing regulators within PcG bodies (46,85). Mechanistically, PcGs often affect chromosome remodeling and compression through PTMs of histones, including methylation of lysine 27 by PRC2 and recruitment of PRC1, to promote chromatin condensation, thereby silencing many important genes. The interaction of PcG protein and cis-regulatory DNA module, termed polycomb response elements (PREs), resulted in remarkable repression of the Hox gene (47). PcG bodies have also been identified as sumoylation centers, resulting from the co-collocation of SUMO E2 (Ubc9) and the substrates (CtBP and CTCF) in PcG bodies, thereby serving as SUMO E3 ligases (86).
Perinucleolar compartment (PNC)
The PNC is known to be enriched with RNA processing proteins and RNA pol III transcripts around the nucleolus (87). Based on these proteins, polypyrimidine tract-binding protein often plays essential roles in pre-mRNA splicing, RNA modification, and translation (88). RNase P and RNase mitochondrial RNA processing (MRP), a subset of RNA pol III transcripts belonging to sequence-specific endoribonucleases, catalyze tRNA maturation and pre-ribosomal RNA processing and mitochondrial DNA replication, respectively (89). Based on the significant progress of PNC, it may facilitate RNA processing and assembly of RNA trafficking complexes, therefore regarded as a transitional separation space in which newly synthesized pol III RNAs and the assembly of their final functional complex are performed. Another important feature of PNC is a unique nuclear body that is closely associated with cell metastatic capacity. Frankowski reported that metarrestin, a PNC inhibitor, effectively suppresses metastatic and invasive behaviors of cancer cells (48).
PML body
PML bodies are ubiquitous MLOs in eukaryotic cells, exerting diverse nuclear functions involved in biological processes associated with DNA replication, gene transcription, or epigenetic gene silencing (90). Kurihara found that PML bodies excluded DNA methyltransferase DNMT3A, which maintained the hypo-methylated state at Y-linked gene promoters, thereby providing a novel insight into the functional properties of nuclear structures in gene transcription (91). PML protein is the essential organizer of PML bodies, of which the protein modified with SUMO to form the ubiquitin-like protein plays a critical role in the recruitment of partners. Dai demonstrated that SUMO-modified PML proteins, termed SUMOylation, promoted the development of liver fibrosis (49). A recent study by Stubbe and colleagues demonstrated that PML bodies were enriched around viral replication centers when viral DNA binding proteins were modified with SUMOs, thereby mediating the replication of the virus (50). In addition, Sehgal revealed that the murine cognate Mx1 could form nuclear condensates overlapping with PML nuclear bodies, further exerting a vital antiviral activity (51).
Histone locus body (HLB)
The HLB is a subnuclear body that frequently appears around the histone-replicating loci to recruit various factors necessary for replication-coupled histone gene transcription and histone pre-mRNAs processing (92). Previous studies demonstrated that HLBs and CBs are not only physically associated but also have the same contents. Coilin, a marker for CBs, was often found in HLBs in high concentrations. It is easy to speculate that the components of HLBs are relatively immobile because they exchange molecules with their environment for a short period, therefore maintaining the stable functioning of metabolically active bodies. Salzler explored the formation mechanism of HLBs using a transgenic assay for ectopic Drosophila HLB assembly and demonstrated that HLB assembly was triggered through a special sequence in the H3-H4 bidirectional promoter, resulting in the expression of histone genes (93).
Paraspeckle
Paraspeckles are one of the essential subnuclear MLOs assembled in the interchromatin space of mammalian cells and play a key role in regulating gene expression (94-96). The majority of paraspeckle proteins harboring prion-like domains undergo LLPS through protein-protein interactions. Previous evidence showed that RBM14 and FUS with an intact prion-like domain effectively constructed paraspeckles (97). Several molecular mechanisms are underlying paraspeckle function. Paraspeckles have been shown to effectively regulate certain RNA species in the nucleus based on the nuclear retention mechanism (98). Besides, paraspeckles rapidly sequester their protein components within a spatial space, resulting in a high concentration in local areas compared to their surroundings in the nuclear, exerting accurate and efficient biological functions. Jiang demonstrated that paraspeckle proteins, SFPQ and NONO, coupled with various miRNA processing factors effectively facilitate miRNA formation (99). Furthermore, paraspeckles can regulate gene expression in various developmental and disease scenarios, including mammalian development, response to viral infection, and tumor process (52).
Nuclear membrane
Nuclear pore complex (NPC)
Several biological processes, including gene expression, chromatin organization, and DNA repair, depend on combined regulation of the nucleus and the cytoplasm, especially for the continuous exchange of various molecules (100). NPCs are assembled into the nuclear envelope through the fusion of the outer and inner nuclear membranes, forming a special channel for molecule transport, to facilitate nucleocytoplasmic transport (101). The principal structural element of NPC includes a five-layer architecture from the nucleus to the cytoplasm as follows: the nuclear basket, the nuclear rings, the inner pore ring, the cytoplasmic rings, and the cytoplasmic filaments (102,103). The NPC structure on the cytoplasmic side, the peripheral elements of cytoplasmic rings, associates with the cytoplasmic filaments that have a flexible property due to the components of intrinsically disordered domains.
Recent studies revealed that NPC facilitates nucleocytoplasmic transport in both directions, including free diffusion for small molecules and major active transport routes for various proteins and RNAs (104). Specifically, NPC-mediated import and export of macromolecules contain three cases, including the export of various nuclear RNAs, the import of ribosomal proteins and transcription factors, and the bi-directional shuttling of molecules associated with various signaling pathways. Aberrant scaffold nucleoporins (NUPs) owing to abnormal expression levels, mutations, and gene fusions, especially for human NUP214, NUP188, and NUP358, have been documented in various human cancers (53). NUP-associated cancer could be interpreted as being caused by significant changes in epigenetic chromatin modification resulting from the fallacious interactions of NUPs with chromatin and abnormal transport of cancer-related factors related to the carcinogenic or tumor-suppression process. A recent study of human Nup210 in the adaptive immune system revealed that Nup210-mediated activation of the TCR signaling is an essential prerequisite for naïve CD4+ T cell homeostasis (54).
Cytoplasm
Processing body (PB)
In eukaryotic cells, PBs are one class of MLOs in the cytosol with multiple biological processes related to translational repression and the mRNA decay machinery (105). PBs have been shown to display a wide array of dynamic behaviors (106). Recent studies revealed the mechanisms underlying the formation of PBs associated with phase transition processes, in which several types of molecular interactions mediating LLPS drive RNAs and proteins to form PBs. Moreover, the PTMs of PB components remarkably affect their formation, including phosphorylation of threonine and serine and methylation of arginine. DCP1A phosphorylated by JNK tends to depart from PBs, while JNK-mediated 4E-T hyperphosphorylation can form the larger PBs (107,108). AKT3-mediated AGO2 phosphorylation significantly enhances the interaction with other proteins, such as TNRC6A/GW182 and DDX6, facilitating PBs formation (109). Matsumoto indicated that RAP55 is important for the assembly of cytoplasmic mRNP granules and PRMT1, a protein arginine methyltransferase, is a prerequisite for RAP55A to localize to PBs (110). One major class of PB compositions is associated with various proteins related to RNA metabolism, further demonstrating that they exert several functions for gene expression. For instance, the XRN1 exonuclease and DCP1/2 decapping factor can control the 5'–3' mRNA decay, while helicase DDX6 and RBP CPEB1 are translational repression factors mediating mRNA translation (111,112). Interestingly, in virus-infected cells, the human interferon (IFN)-inducible ‘myxovirus resistance protein A (MxA)’ displays antiviral activity with P bodies containing respective viral nucleocapsid proteins (55).
Uridine-rich snRNPs (U snRNPs) body (U body)
U snRNPs play essential roles in pre-mRNA processing in the nucleus. However, recent studies revealed that they are assembled in the cytoplasm of eukaryotic cells (113). Generally, cytoplasmic SMN protein-containing granules, termed U bodies, have a main component associated with U snRNPs, which is coupled with various essential snRNP assembly factors. FRAP assays have indicated that U bodies are spherical, which often has a characteristic on significant dynamics, including fusion or fission of bodies (114). Tsalikis reported that U body formation was associated with the induction of metabolic stresses, such as amino acid starvation, which was triggered by cell membrane damage due to infection with intracellular bacterial pathogens, while U bodies significantly disappeared once the stress was removed, indicating that the process of U body assembly would characterize an adaptive cell-response to metabolic stress (56).
Balbiani body (Bb)
The Bb granule is a non-membrane bound and a dynamic compartment that assembles early during oocyte formation and disappears in late-stage oocytes in mammals (115). Boke showed that Xvelo harboring an N-terminal prion-like domain can form Xenopus Bbs, accompanied by co-recruitment of mitochondria and RNA (116). Escobar-Aguirre showed that microtubule-actin crosslinking factor 1α (macf1α) possesses an essential role in regulating Bb disassembly and oocyte nucleus positioning, which was demonstrated using CRISPR/Cas9 genome editing technology to delete these domains by targeting macf1α endogenous gene (117). Kaufman revealed that the wild-type rbpms2 protein was closely associated with spherical Bb formation, while Bbs disappeared in rbpms2 mutant oocytes, suggesting the crucial role of wild-type rbpms2 protein in Bb assembly (118).
Germ granule
Germ granules are MLOs associated with RNA-rich cytoplasmic bodies of germ cells, carrying many proteins and RNAs related to post-transcriptional regulation specific to germ cells (119,120). A study by Brangwynne revealed that individual components involved in germ granules are highly dynamic (16). Germ granules play significant roles in the development of germ cells. First, germ granule has been implicated in post-transcriptional regulation of mRNAs, such as mRNA localization, stability, and translational activity (121). Seydoux showed that germ granules may be essential to preserve the plasticity of the germline genome due to the control of gene expression in germ cells (57). Second, germ granule determines the germ cell fate because germ granules are used as a part of the germplasm.
Transport RNP
Transport RNPs are prevalent in neurons, and often harbor different mRNAs and associated binding proteins, such as elongation factors and ribosomal proteins or even clusters of ribosomes, possibly serving as subunits of molecular motors that are essential for mRNA transport (122). Monani showed that SMN and Gemin proteins involved in the RNP complex had a beneficial effect on the assembly of spliceosomal RNPs, further playing an essential role in the formation of transport RNPs (123). Additionally, previous studies demonstrated that transport RNPs could facilitate RNA localization and achieve precise translational control.
Synaptic density
In addition to MOs and MLOs, there is a unique type of membrane-semi-enclosed compartments in neuron cells, termed synapses, that mediate signal processing and transmission involved in the nervous system (124). Postsynaptic densities (PSDs) are typical synapses that contain glutamate receptors and associated signaling and structural molecules. Zeng reported that the multivalent postsynaptic protein-protein interaction between SynGAP and PSD-95 can contribute to the LLPS process, which is possibly a novel mechanism for PSD formation (58). Recent studies indicated that molecular components within PSDs could exert highly dynamic activity, constantly exchanging their components with bulk aqueous cytoplasm in synaptic spines (125). Aberrant development and regulation of PSD often result in an imbalance between excitation and inhibition in neuronal circuits, which causes multiple diseases, such as autism, intellectual disability, and mental illness.
Stress granule (SG)
SG represents assemblies of untranslated messenger RNPs (mRNPs) derived from mRNAs arrested in translation initiation (126,127). Interestingly, SGs have two distinct layers, including a core structure with higher concentrations of proteins and mRNAs and a potentially dynamic shell (128). FRAP assays have shown that most components of SGs are rapidly exchanged, thereby undergoing fusion and fission in the cytosol, suggesting that SGs are dynamic structures (129). A dense network of protein-protein interactions between mRNA-binding proteins significantly contributes to SG formation, which is modulated by various PTMs, including methylation, phosphorylation, and glycosylation. Goulet demonstrated that methylated arginine is necessary for the recruitment of TDRD3 in SGs, facilitating the protein aggregation in SGs (130). Several observations showed that phosphorylated protein impairs granule assembly, suggesting that SG disassembly is promoted by Grb7 phosphorylation and DYRK3 kinase in focal adhesion kinase (FAK) and mTORC1 signaling pathway, respectively (131,132). In addition, Ohn found that O-Glc-NAc glycosylation of proteins enhances SG formation and Duan suggested a crucial role of PARylation in regulating the dynamics of RNP granules, contributing to ALS disease pathogenesis by promoting SG formation (59,60).
SG formation plays an essential role in several biological processes. First, SGs rapidly recruit numerous antiviral proteins at a high local concentration after viral infection, further enhancing the activation of innate immune response and improving viral resistance (133,134). Second, SGs have been proposed to modulate signaling pathways by sequestering components of TOR, RACK1, or TRAF2 signaling pathways (135,136). Importantly, SG formation often seems to be involved in various human diseases, including several degenerative diseases such as ALS, frontotemporal lobar degeneration (FTLD), and various myopathies (137). In addition, SG formation contributes to tumor progression, and chemotherapy drugs effectively disturb SG formation, which might provide a new strategy for cancer treatment.
Plasma membrane
Immune synapse
An immune synapse is often formed on the surface of activated immune cells, such as T cells, B cells, and natural killer (NK) cells (138). Upon encounter with associated antigens, immune cells are activated by the reorganization of its membrane, rapidly forming a specific compartment known as the immune synapse. Benard et al. found that LFA-1 and CD28 molecules play a crucial role in enhancing the impact of TCR clustering on cell spreading and actin organization (139,140). In addition, Nowosad and colleagues have demonstrated that germinal center B cells recognize antigens through a specialized immune synapse architecture, thus B cells can selectively recruit antigens by altering individual components in the synaptic architecture (141). For NK cells, recent data have revealed that NKp46 signaling directly regulates the associated immune synapse rearrangement and several immune synapse-related functions (142).
Focal adhesions (FAs)
FA is a special non-membrane structure composed of clustered transmembrane proteins and signaling proteins, such as integrin, talin, paxillin, vinculin, and FAK (143). Recent studies demonstrated that FAs often link to the cellular cytoskeleton, further initiating cell activities, such as cell migration, proliferation, and differentiation (144). Cho reported that the FA molecule, matrix metalloproteinase 1 (Mmp1), effectively regulates astrocyte morphology and glutamate transporters to suppress seizure-like behavior (145). Png showed that transglutaminase-2 influenced phosphorylation of paxillin by JNK, further interacting with matrix proteins and integrins, thereby forming FAs (146). Luo found that ARAP2, an Arf GTPase-activating protein, affects FA dynamics in an Akt activity-dependent manner, further regulating the size and number of FAs (147). Toro-Tapia proposed that Ric-8A-mediated Gα13 signaling is required for proper cell migration by controlling FA dynamics (148).
Podosome
Podosomes are multimolecular cytoskeletal structures that are particularly formed in various monocytic lineage cells, endothelial cells, and smooth muscle cells (149). Interestingly, podosomes can achieve the cell-matrix linkage between cells and their surroundings, such as integrins (150). Therefore, these dynamic interactions suggest that podosomes may continuously recognize the surrounding information, further reshaping the pericellular environment. Previous studies highlighted the specific functions of podosomes in several primary human monocytic cells, including cell adhesion, cell extravasation and invasion, cell membrane fusion, and tissue remodeling (151).
Conclusions
Given that various biomolecules can undergo the LLPS process under idealized conditions, there is great interest in understanding the specific properties of biological condensates. Many previous studies suggested that biomolecular condensates have pervasive roles in cell biology. However, research in this field tends to progress from the phase behavior of biomolecules to a general platform that explains and predicts bio-functional effects of these biomolecules. Several trials with cellular extracts are essential approaches that may help to explore the functional effects of biomolecular condensates in cellular processes. In this review, we summarize and highlight the diverse biological functions of condensates, providing an important framework for a better understanding of the biological processes associated with the phase behavior of biomolecules (Table 1).
Table 1
| Location | MLO name | Defining components | Functions | Diseases |
|---|---|---|---|---|
| Nucleus | Cajal body | Coilin, SMN | Pre-mRNA splicing, pre-rRNA processing | ALS, SMA (43) |
| Cleavage body | CstF-64, CPSF-100 | DNA replication, processing | NA | |
| Gem | SMN | Pre-mRNA splicing, RNA metabolism | SMA (44) | |
| Nuclear speckle | snRNPs, SR proteins, Malat1 | Gene transcription, RNA processing | NA | |
| Nucleolus | RNA Pol I machinery | rDNA transcription, pre-rRNA processing, ribosome biogenesis | Werner syndrome, Treacher Collins syndrome, dyskeratosis congenita syndrome, Rothmund-Thomson syndrome (45) | |
| OPT domain body | PTF, Oct1, TBF, Sp1 | Gene transcription, antiviral defense | NA | |
| PcG body | PRC1, PRC2, PREs | Transcriptional repression, sumoylation centers | Cancer (46,47) | |
| Perinucleolar compartment | CUGBP, KSRP | tRNA maturation, pre-ribosomal RNA processing, mitochondrial DNA replication | Cancer (48) | |
| PML body | PML, DNMT3A, Mx1 | DNA replication, transcription, epigenetic gene silencing | Liver fibrosis (49), antiviral defense (50,51) | |
| Histone locus body | NPAT, FLASH | Histone transcription, pre-mRNAs processing | NA | |
| Paraspeckle | CTN-RNA, PSP1, p54nrb | RNA processing | Mammalian development, antiviral defense, cancer (52) | |
| Nuclear membrane | Nuclear pore complex | FG-NUPs | Nucleocytoplasmic transport | Cancer (53), immune (54) |
| Cytoplasm | P body | DCP1A, TNRC6A/GW18, DDX6, RAP55, MxA | mRNA metabolism | (55) |
| U body | SMN | snRNP metabolism | SMA (56) | |
| Balbiani Body | macf1α, rbpms2 | RNA storage | NA | |
| Germ granule | NA | Post-transcriptional regulation | Germ cell development (57) | |
| Transport RNP | Staufen1, Staufen2, FMRP, ZBP1, hnRNPA2, CPEB, Pura, and SMN | mRNA transport | NA | |
| Synaptic density | SynGAP, PSD-95 | Signal processing and transmission | Autism, intellectual disability, mental illness (58) | |
| Stress granule | RBPs, non-RBPs, TDRD3 | Translational regulation, mRNA storage | ALS, FTLD, some myopathies (59,60) | |
| Plasma membrane | Immune synapse | TCR, NKp46 | Immune synapse rearrangement | NA |
| Focal adhesions | Integrin, talin, paxillin, vinculin, FAK, Mmp1 | Cell migration, proliferation, differentiation | NA | |
| Podosome | actin-rich proteins, actin-associated proteins | Cell adhesion, migration | NA |
MLO, membraneless organelle, NA, not applicable; OPT, Oct1/PTF/transcription; PcG, polycomb group; PML, promyelocytic leukemia; P body, processing body; U body, uridine-rich snRNPs body; ALS, amyotrophic lateral sclerosis; SMA, spinal muscular atrophy; FTLD, frontotemporal lobar degeneration.
First, intracellular MLOs may induce some adaptive and reversible responses that are exquisitely sensitive to changes in physicochemical conditions involved in the extracellular environment, including alteration of transcriptional or translational processes (152,153). In addition, MLOs are essential in controlling endogenous cellular activities through sequestration of molecules; therefore, some components are recruited into a dense phase whereas others distribute in the dilute phase, thereby inhibiting multiple biological processes, such as enzymatic reaction or signaling transduction. Second, MLOs can be used to control biomolecule concentrations in living cells. When the concentration of a biomolecule is in a saturation state, phase-separated MLOs can be locally formed. Interestingly, excess protein stored in organelles with high concentrations generally creates a replenishment pool for biomolecules in cells; intramolecules spontaneously enter the surrounding environment to promote biological processes when the concentration of biomolecules drops. In addition, LLPS driving high concentration of molecules in condensates may activate and accelerate biochemical reactions, promoting various signaling processes. Third, MLOs formed by LLPS may be important for maintaining cell morphogenesis. Specifically, LLPS can mediate the formation of materials with viscoelastic properties, improving cellular structures, including many membrane- and membraneless-containing organelles (154). Schmidt et al. demonstrated that LLPS formed by dynamical interactions between the macromolecules facilitates the formation of nuclear pores (155).
In 2019, a novel coronavirus, termed SARS-CoV-2, has rapidly spread around the world, leading to a new public health concern. Interestingly, previous studies revealed that the life cycle of SARS-CoV-2 is closely related to the LLPS phenomenon, including SARS-CoV-2 assembly and replication. For instance, Chen reported that the LLPS of nucleocapsid (N) protein and genome RNA may be essential driving force for SARS-CoV-2 viral assembly, highlighting intervention strategies to combat SARS-CoV-2 infections through the impaired viral assembly attributed to disrupting the LLPS (156). Iserman demonstrated that the nucleocapsid protein (N-protein) may rapidly undergo LLPS. In particular, RNA sequences located at 5' and 3’ ends of the genome may promote N-protein condensation while other genomic regions (frameshifting region) exert a special function on facilitating condensate dissolution, presenting a identifying platform for detecting antiviral compounds effective against SARS-CoV-2 based on targeting phase-separation (157). Savastano suggested the RNA genome may effectively induce nucleocapsid protein LLPS, resulting in generation of high-density biomolecule condensates containing viral RNA-dependent RNA polymerase that determines the SARS-CoV-2 replication capacity, and finally providing a theoretical basis for the design of novel therapeutics to combat SARS-CoV-2 through inhibition of the RNA-induced phase separation of N protein (158).
MLOs produced by LLPS in cells may concentrate biomolecules (proteins and nucleic acids) with high concentrations compared to the surrounding environment. This phenomenon has essential functions in multiple cellular processes, such as RNA metabolism, regulation of gene expression, stress adaptation, and transmembrane signaling cascades. The present review, evaluated the current state of knowledge of MLOs, highlighting their biogenesis, organization, dynamics, and function. Understanding phase separation could pave the way for developing novel therapeutic strategies for the treatment of LLPS-related diseases or tumors in the future.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1111
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1111). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Klosin A, Oltsch F, Harmon T, et al. Phase separation provides a mechanism to reduce noise in cells. Science 2020;367:464-8. [Crossref] [PubMed]
- Ong JY, Torres JZ. Phase Separation in Cell Division. Mol Cell 2020;80:9-20. [Crossref] [PubMed]
- Liu Z, Zhang S, Gu J, et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat Struct Mol Biol 2020;27:363-72. [Crossref] [PubMed]
- Fuller GG, Han T, Freeberg MA, et al. RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. Elife 2020;9:48480. [Crossref] [PubMed]
- Adame-Arana O, Weber CA, Zaburdaev V, et al. Liquid Phase Separation Controlled by pH. Biophys J 2020;119:1590-605. [Crossref] [PubMed]
- Zhao YG, Zhang H. Phase Separation in Membrane Biology: The Interplay between Membrane-Bound Organelles and Membraneless Condensates. Dev Cell 2020;55:30-44. [Crossref] [PubMed]
- Woodruff JB, Hyman AA, Boke E. Organization and Function of Non-dynamic Biomolecular Condensates. Trends Biochem Sci 2018;43:81-94. [Crossref] [PubMed]
- Boeynaems S, Alberti S, Fawzi NL, et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 2018;28:420-35. [Crossref] [PubMed]
- Banani SF, Lee HO, Hyman AA, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017;18:285-98. [Crossref] [PubMed]
- Sabari BR, Dall'Agnese A, Boija A, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018;361:eaar3958. [Crossref] [PubMed]
- Kaganovich D. There Is an Inclusion for That: Material Properties of Protein Granules Provide a Platform for Building Diverse Cellular Functions. Trends Biochem Sci 2017;42:765-76. [Crossref] [PubMed]
- Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell 2013;153:287-92. [Crossref] [PubMed]
- Yasuda S, Tsuchiya H, Kaiho A, et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020;578:296-300. [Crossref] [PubMed]
- Su X, Ditlev JA, Hui E, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016;352:595-9. [Crossref] [PubMed]
- Riback JA, Zhu L, Ferrolino MC, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 2020;581:209-14. [Crossref] [PubMed]
- Brangwynne CP, Eckmann CR, Courson DS, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009;324:1729-32. [Crossref] [PubMed]
- Sanders DW, Kedersha N, Lee DSW, et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020;181:306-324.e28. [Crossref] [PubMed]
- Lin YH, Forman-Kay JD, Chan HS. Theories for Sequence-Dependent Phase Behaviors of Biomolecular Condensates. Biochemistry 2018;57:2499-508. [Crossref] [PubMed]
- Wang J, Choi JM, Holehouse AS, et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018;174:688-699.e16. [Crossref] [PubMed]
- Li P, Banjade S, Cheng HC, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012;483:336-40. [Crossref] [PubMed]
- Banani SF, Rice AM, Peeples WB, et al. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016;166:651-63. [Crossref] [PubMed]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007;8:1006-16. [Crossref] [PubMed]
- Pak CW, Kosno M, Holehouse AS, et al. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell 2016;63:72-85. [Crossref] [PubMed]
- Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012;149:753-67. [Crossref] [PubMed]
- Maharana S, Wang J, Papadopoulos DK, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018;360:918-21. [Crossref] [PubMed]
- Conicella AE, Dignon GL, Zerze GH, et al. TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proc Natl Acad Sci U S A 2020;117:5883-94. [Crossref] [PubMed]
- Xiang S, Kato M, Wu LC, et al. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 2015;163:829-39. [Crossref] [PubMed]
- Söding J, Zwicker D, Sohrabi-Jahromi S, et al. Mechanisms for Active Regulation of Biomolecular Condensates. Trends Cell Biol 2020;30:4-14. [Crossref] [PubMed]
- Kholodenko BN, Hoek JB, Westerhoff HV. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol 2000;10:173-8. [Crossref] [PubMed]
- Ditlev JA, Vega AR, Köster DV, et al. A composition-dependent molecular clutch between T cell signaling condensates and actin. Elife 2019;8:42695. [Crossref] [PubMed]
- Houtman JC, Yamaguchi H, Barda-Saad M, et al. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat Struct Mol Biol 2006;13:798-805. [Crossref] [PubMed]
- Snead WT, Gladfelter AS. The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, PTMs, and Active Processes Regulate Condensation. Mol Cell 2019;76:295-305. [Crossref] [PubMed]
- Monahan Z, Ryan VH, Janke AM, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J 2017;36:2951-67. [Crossref] [PubMed]
- Guillén-Boixet J, Buzon V, Salvatella X, et al. CPEB4 is regulated during cell cycle by ERK2/Cdk1-mediated phosphorylation and its assembly into liquid-like droplets. Elife 2016;5:19298. [Crossref] [PubMed]
- Wang A, Conicella AE, Schmidt HB, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J 2018;37:e97452. [Crossref] [PubMed]
- Qamar S, Wang G, Randle SJ, et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018;173:720-734.e15. [Crossref] [PubMed]
- Ryan VH, Dignon GL, Zerze GH, et al. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol Cell 2018;69:465-479.e7. [Crossref] [PubMed]
- Saito M, Hess D, Eglinger J, et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol 2019;15:51-61. [Crossref] [PubMed]
- Cohen TJ, Hwang AW, Restrepo CR, et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nat Commun 2015;6:5845. [Crossref] [PubMed]
- Yuzwa SA, Shan X, Macauley MS, et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol 2012;8:393-9. [Crossref] [PubMed]
- Roth S, Khalaila I. The effect of O-GlcNAcylation on hnRNP A1 translocation and interaction with transportin1. Exp Cell Res 2017;350:210-7. [Crossref] [PubMed]
- Marotta NP, Lin YH, Lewis YE, et al. O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson's disease. Nat Chem 2015;7:913-20. [Crossref] [PubMed]
- Zhang Z, Lotti F, Dittmar K, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008;133:585-600. [Crossref] [PubMed]
- Groen EJN, Talbot K, Gillingwater TH. Advances in therapy for spinal muscular atrophy: promises and challenges. Nat Rev Neurol 2018;14:214-24. [Crossref] [PubMed]
- Pederson T. The nucleolus. Cold Spring Harb Perspect Biol 2011;3:a000638. [Crossref] [PubMed]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011;469:343-9. [Crossref] [PubMed]
- Bantignies F, Roure V, Comet I, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011;144:214-26. [Crossref] [PubMed]
- Frankowski KJ, Wang C, Patnaik S, et al. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci Transl Med 2018;10:eaap8307. [Crossref] [PubMed]
- Dai J, Hu Y, Niu Q, et al. Role of PML SUMOylation in arsenic trioxide-induced fibrosis in HSCs. Life Sci 2020;251:117607. [Crossref] [PubMed]
- Stubbe M, Mai J, Paulus C, et al. Viral DNA Binding Protein SUMOylation Promotes PML Nuclear Body Localization Next to Viral Replication Centers. mBio 2020;11:00049-20. [Crossref] [PubMed]
- Sehgal PB, Yuan H, Scott MF, et al. Murine GFP-Mx1 forms nuclear condensates and associates with cytoplasmic intermediate filaments: Novel antiviral activity against VSV. J Biol Chem 2020;295:18023-35. [Crossref] [PubMed]
- Wang Y, Hu SB, Wang MR, et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol 2018;20:1145-58. [Crossref] [PubMed]
- Gozalo A, Duke A, Lan Y, et al. Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression. Mol Cell 2020;77:67-81.e7. [Crossref] [PubMed]
- Borlido J, Sakuma S, Raices M, et al. Nuclear pore complex-mediated modulation of TCR signaling is required for naïve CD4+ T cell homeostasis. Nat Immunol 2018;19:594-605. [Crossref] [PubMed]
- Sehgal PB, Westley J, Lerea KM, et al. Biomolecular condensates in cell biology and virology: Phase-separated membraneless organelles (MLOs). Anal Biochem 2020;597:113691. [Crossref] [PubMed]
- Tsalikis J, Tattoli I, Ling A, et al. Intracellular Bacterial Pathogens Trigger the Formation of U Small Nuclear RNA Bodies (U Bodies) through Metabolic Stress Induction. J Biol Chem 2015;290:20904-18. [Crossref] [PubMed]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell 2006;127:891-904. [Crossref] [PubMed]
- Zeng M, Chen X, Guan D, et al. Reconstituted Postsynaptic Density as a Molecular Platform for Understanding Synapse Formation and Plasticity. Cell 2018;174:1172-1187.e16. [Crossref] [PubMed]
- Ohn T, Kedersha N, Hickman T, et al. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 2008;10:1224-31. [Crossref] [PubMed]
- Duan Y, Du A, Gu J, et al. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res 2019;29:233-47. [Crossref] [PubMed]
- Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol 2003;4:975-80. [Crossref] [PubMed]
- Staněk D. Cajal bodies and snRNPs - friends with benefits. RNA Biol 2017;14:671-9. [Crossref] [PubMed]
- Lafarga V, Tapia O, Sharma S, et al. CBP-mediated SMN acetylation modulates Cajal body biogenesis and the cytoplasmic targeting of SMN. Cell Mol Life Sci 2018;75:527-46. [Crossref] [PubMed]
- Girard C, Neel H, Bertrand E, et al. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res 2006;34:2925-32. [Crossref] [PubMed]
- Platani M, Goldberg I, Swedlow JR, et al. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 2000;151:1561-74. [Crossref] [PubMed]
- Pellizzoni L, Charroux B, Rappsilber J, et al. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol 2001;152:75-85. [Crossref] [PubMed]
- Schul W, Groenhout B, Koberna K, et al. The RNA 3' cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J 1996;15:2883-92. [Crossref] [PubMed]
- Li L, Roy K, Katyal S, et al. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol Biol Cell 2006;17:1126-40. [Crossref] [PubMed]
- Feng W, Gubitz AK, Wan L, et al. Gemins modulate the expression and activity of the SMN complex. Hum Mol Genet 2005;14:1605-11. [Crossref] [PubMed]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011;13:167-73. [Crossref] [PubMed]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol 2011;3:a000646. [Crossref] [PubMed]
- Tsaytler P, Harding HP, Ron D, et al. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 2011;332:91-4. [Crossref] [PubMed]
- Melcák I, Cermanová S, Jirsová K, et al. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol Biol Cell 2000;11:497-510. [Crossref] [PubMed]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature 1997;387:523-7. [Crossref] [PubMed]
- Boisvert FM, van Koningsbruggen S, Navascués J, et al. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007;8:574-85. [Crossref] [PubMed]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol 2006;16:19-26. [Crossref] [PubMed]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet 2011;27:295-306. [Crossref] [PubMed]
- Dimario PJ. Cell and molecular biology of nucleolar assembly and disassembly. Int Rev Cytol 2004;239:99-178. [Crossref] [PubMed]
- Visintin R, Amon A. The nucleolus: the magician's hat for cell cycle tricks. Curr Opin Cell Biol 2000;12:372-7. [Crossref] [PubMed]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 2003;22:6068-77. [Crossref] [PubMed]
- Dove BK, You JH, Reed ML, et al. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell Microbiol 2006;8:1147-57. [Crossref] [PubMed]
- Harrigan JA, Belotserkovskaya R, Coates J, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 2011;193:97-108. [Crossref] [PubMed]
- Pombo A, Cuello P, Schul W, et al. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J 1998;17:1768-78. [Crossref] [PubMed]
- Minezaki Y, Homma K, Kinjo AR, et al. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol 2006;359:1137-49. [Crossref] [PubMed]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009;10:697-708. [Crossref] [PubMed]
- Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell 2003;113:127-37. [Crossref] [PubMed]
- Pollock C, Huang S. The perinucleolar compartment. J Cell Biochem 2009;107:189-93. [Crossref] [PubMed]
- Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 2004;23:3200-7. [Crossref] [PubMed]
- van Eenennaam H, Jarrous N, van Venrooij WJ, et al. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life 2000;49:265-72. [Crossref] [PubMed]
- Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010;2:a000661. [Crossref] [PubMed]
- Kurihara M, Kato K, Sanbo C, et al. Genomic Profiling by ALaP-Seq Reveals Transcriptional Regulation by PML Bodies through DNMT3A Exclusion. Mol Cell 2020;78:493-505.e8. [Crossref] [PubMed]
- Wagner EJ, Burch BD, Godfrey AC, et al. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 2007;28:692-9. [Crossref] [PubMed]
- Salzler HR, Tatomer DC, Malek PY, et al. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev Cell 2013;24:623-34. [Crossref] [PubMed]
- Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol 2013;10:456-61. [Crossref] [PubMed]
- Prasanth KV, Prasanth SG, Xuan Z, et al. Regulating gene expression through RNA nuclear retention. Cell 2005;123:249-63. [Crossref] [PubMed]
- Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33:717-26. [Crossref] [PubMed]
- Hennig S, Kong G, Mannen T, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015;210:529-39. [Crossref] [PubMed]
- Torres M, Becquet D, Blanchard MP, et al. Circadian RNA expression elicited by 3'-UTR IRAlu-paraspeckle associated elements. Elife 2016;5:14837. [Crossref] [PubMed]
- Jiang L, Shao C, Wu QJ, et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol 2017;24:816-24. [Crossref] [PubMed]
- D'Angelo MA. Nuclear pore complexes as hubs for gene regulation. Nucleus 2018;9:142-8. [Crossref] [PubMed]
- Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol 2017;18:229-45. [Crossref] [PubMed]
- Hampoelz B, Andres-Pons A, Kastritis P, et al. Structure and Assembly of the Nuclear Pore Complex. Annu Rev Biophys 2019;48:515-36. [Crossref] [PubMed]
- Amlacher S, Sarges P, Flemming D, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 2011;146:277-89. [Crossref] [PubMed]
- Sarma A, Yang W. Calcium regulation of nucleocytoplasmic transport. Protein Cell 2011;2:291-302. [Crossref] [PubMed]
- Riggs CL, Kedersha N, Ivanov P, et al. Mammalian stress granules and P bodies at a glance. J Cell Sci 2020;133:jcs242487. [Crossref] [PubMed]
- Aizer A, Brody Y, Ler LW, et al. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell 2008;19:4154-66. [Crossref] [PubMed]
- Rzeczkowski K, Beuerlein K, Müller H, et al. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol 2011;194:581-96. [Crossref] [PubMed]
- Cargnello M, Tcherkezian J, Dorn JF, et al. Phosphorylation of the eukaryotic translation initiation factor 4E-transporter (4E-T) by c-Jun N-terminal kinase promotes stress-dependent P-body assembly. Mol Cell Biol 2012;32:4572-84. [Crossref] [PubMed]
- Horman SR, Janas MM, Litterst C, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell 2013;50:356-67. [Crossref] [PubMed]
- Matsumoto K, Nakayama H, Yoshimura M, et al. PRMT1 is required for RAP55 to localize to processing bodies. RNA Biol 2012;9:610-23. [Crossref] [PubMed]
- van Dijk E, Cougot N, Meyer S, et al. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 2002;21:6915-24. [Crossref] [PubMed]
- Wilczynska A, Aigueperse C, Kress M, et al. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 2005;118:981-92. [Crossref] [PubMed]
- Suzuki T, Izumi H, Ohno M. Cajal body surveillance of U snRNA export complex assembly. J Cell Biol 2010;190:603-12. [Crossref] [PubMed]
- Buckingham M, Liu JL. U bodies respond to nutrient stress in Drosophila. Exp Cell Res 2011;317:2835-44. [Crossref] [PubMed]
- Bilinski SM, Kloc M, Tworzydlo W. Selection of mitochondria in female germline cells: is Balbiani body implicated in this process? J Assist Reprod Genet 2017;34:1405-12. [Crossref] [PubMed]
- Boke E, Ruer M, Wühr M, et al. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016;166:637-50. [Crossref] [PubMed]
- Escobar-Aguirre M, Zhang H, Jamieson-Lucy A, et al. Microtubule-actin crosslinking factor 1 (Macf1) domain function in Balbiani body dissociation and nuclear positioning. PLoS Genet 2017;13:e1006983. [Crossref] [PubMed]
- Kaufman OH, Lee K, Martin M, et al. Correction: rbpms2 functions in Balbiani body architecture and ovary fate. PLoS Genet 2018;14:e1007768. [Crossref] [PubMed]
- Voronina E, Seydoux G, Sassone-Corsi P, et al. RNA granules in germ cells. Cold Spring Harb Perspect Biol 2011;3:a002774. [Crossref] [PubMed]
- Trcek T, Grosch M, York A, et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun 2015;6:7962. [Crossref] [PubMed]
- Lehtiniemi T, Kotaja N. Germ granule-mediated RNA regulation in male germ cells. Reproduction 2018;155:R77-91. [Crossref] [PubMed]
- Tang SJ, Meulemans D, Vazquez L, et al. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron 2001;32:463-75. [Crossref] [PubMed]
- Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron 2005;48:885-96. [Crossref] [PubMed]
- Milovanovic D, Wu Y, Bian X, et al. A liquid phase of synapsin and lipid vesicles. Science 2018;361:604-7. [Crossref] [PubMed]
- Milovanovic D, De Camilli P. Synaptic Vesicle Clusters at Synapses: A Distinct Liquid Phase? Neuron 2017;93:995-1002. [Crossref] [PubMed]
- Youn JY, Dyakov BJA, Zhang J, et al. Properties of Stress Granule and P-Body Proteomes. Mol Cell 2019;76:286-94. [Crossref] [PubMed]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 2009;10:430-6. [Crossref] [PubMed]
- Jain S, Wheeler JR, Walters RW, et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016;164:487-98. [Crossref] [PubMed]
- Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol 2016;26:668-79. [Crossref] [PubMed]
- Goulet I, Boisvenue S, Mokas S, et al. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet 2008;17:3055-74. [Crossref] [PubMed]
- Tsai NP, Ho PC, Wei LN. Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO J 2008;27:715-26. [Crossref] [PubMed]
- Wippich F, Bodenmiller B, Trajkovska MG, et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013;152:791-805. [Crossref] [PubMed]
- Reineke LC, Lloyd RE. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol 2015;89:2575-89. [Crossref] [PubMed]
- McCormick C, Khaperskyy DA. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 2017;17:647-60. [Crossref] [PubMed]
- Thedieck K, Holzwarth B, Prentzell MT, et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 2013;154:859-74. [Crossref] [PubMed]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, et al. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 2008;10:1324-32. [Crossref] [PubMed]
- Chen L, Liu B. Relationships between Stress Granules, Oxidative Stress, and Neurodegenerative Diseases. Oxid Med Cell Longev 2017;2017:1809592. [Crossref] [PubMed]
- Dustin ML, Groves JT. Receptor signaling clusters in the immune synapse. Annu Rev Biophys 2012;41:543-56. [Crossref] [PubMed]
- Beemiller P, Krummel MF. Mediation of T-cell activation by actin meshworks. Cold Spring Harb Perspect Biol 2010;2:a002444. [Crossref] [PubMed]
- Benard E, Nunès JA, Limozin L, et al. T Cells on Engineered Substrates: The Impact of TCR Clustering Is Enhanced by LFA-1 Engagement. Front Immunol 2018;9:2085. [Crossref] [PubMed]
- Nowosad CR, Spillane KM, Tolar P. Germinal center B cells recognize antigen through a specialized immune synapse architecture. Nat Immunol 2016;17:870-7. [Crossref] [PubMed]
- Hadad U, Thauland TJ, Martinez OM, et al. NKp46 Clusters at the Immune Synapse and Regulates NK Cell Polarization. Front Immunol 2015;6:495. [Crossref] [PubMed]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol 2015;17:955-63. [Crossref] [PubMed]
- Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 2010;11:633-43. [Crossref] [PubMed]
- Cho S, Muthukumar AK, Stork T, et al. Focal adhesion molecules regulate astrocyte morphology and glutamate transporters to suppress seizure-like behavior. Proc Natl Acad Sci U S A 2018;115:11316-21. [Crossref] [PubMed]
- Png E, Hou A, Tong L. Mechanistic role of transglutaminase-2 in focal adhesions. Sci Rep 2018;8:12370. [Crossref] [PubMed]
- Luo R, Chen PW, Kuo JC, et al. ARAP2 inhibits Akt independently of its effects on focal adhesions. Biol Cell 2018;110:257-70. [Crossref] [PubMed]
- Toro-Tapia G, Villaseca S, Beyer A, et al. The Ric-8A/Gα13/FAK signalling cascade controls focal adhesion formation during neural crest cell migration in Xenopus. Development 2018;145:dev164269. [Crossref] [PubMed]
- Linder S. Invadosomes at a glance. J Cell Sci 2009;122:3009-13. [Crossref] [PubMed]
- Linder S, Kopp P. Podosomes at a glance. J Cell Sci 2005;118:2079-82. [Crossref] [PubMed]
- Cougoule C, Van Goethem E, Le Cabec V, et al. Blood leukocytes and macrophages of various phenotypes have distinct abilities to form podosomes and to migrate in 3D environments. Eur J Cell Biol 2012;91:938-49. [Crossref] [PubMed]
- Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019;176:419-34. [Crossref] [PubMed]
- Dignon GL, Best RB, Mittal J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu Rev Phys Chem 2020;71:53-75. [Crossref] [PubMed]
- Bergeron-Sandoval LP, Safaee N, Michnick SW. Mechanisms and Consequences of Macromolecular Phase Separation. Cell 2016;165:1067-79. [Crossref] [PubMed]
- Schmidt HB, Görlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci 2016;41:46-61. [Crossref] [PubMed]
- Chen H, Cui Y, Han X, et al. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res 2020;30:1143-5. [Crossref] [PubMed]
- Iserman C, Roden CA, Boerneke MA, et al. Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Mol Cell 2020;80:1078-1091.e6. [Crossref] [PubMed]
- Savastano A, Ibáñez de Opakua A, Rankovic M, et al. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun 2020;11:6041. [Crossref] [PubMed]


