
The molecular mechanism of long non-coding ribonucleic acid (lncRNA) RUNX1-IT1 promotes the proliferation and stemness of lung cancer cells
Introduction
Lung cancer is one of the most rapidly increasing malignant tumors in terms of morbidity and death, and represents the largest threat to people’s health and lives (1). Lung cancer is broadly classified into two categories: non-small lung cancer and small cell lung cancer. Non-small cell lung cancer accounts for more than 85% of all lung cancers (2). Conventional treatment approaches for lung cancer include chemotherapy, surgical removal, and radiotherapy; however, due to the metastasis of cancer cells and the low sensitivity of chemotherapy, the treatment effect is not ideal for advanced lung cancer, and the survival rate of patients suffering from lung cancer of about 5 years is unsatisfactory (3,4).
Years of research have proven that lung cancer is caused by the aberrant expression of multiple oncogenes and the deactivation of tumor suppressor genes. The identification of these variations and the revelation of their functions in lung cancer has accelerated the development of targeted therapies for lung cancer, such as the targeting of the small molecule inhibitors of epidermal growth factor receptor, v-raf murine sarcoma viral oncogene homolog B1, anaplastic lymphoma kinase, which have massively enhanced the survival time of lung cancer patients (5,6). The exploration of molecular mechanisms related to lung cancer occurrence and metastasis leads to the development of new concepts and intervention strategies, which is of great significance in improving the clinical efficacy of lung cancer treatments.
Over recent decades, numerous studies have shown that in addition to the above-mentioned related genes that perform important tasks in the onset and advancement of lung cancer, some long-neglected ribonucleic acids (RNAs) without any coding ability, such as non-coding RNA, also play an important role (7). Certain RNA molecules [i.e., long non-coding RNAs (lncRNAs)], with a length of 200 nt–100 kb, are transcribed from cell genomes, which do not have the ability to protein code (8). The abnormal expression of lncRNA may affect the abnormal activation or the inhibition of related gene expression networks in the cells of lung cancer, and then regulate the proliferation, invasion, stemness, and drug resistance of tumor cells. Compared to normal tissues and lung epithelial cells, lncRNA-activated by transforming growth factor β (lncRNA-ATB) is highly expressed in the cancerous cell lines and tissues of the lung, and the overexpression of lncRNA-ATB has been shown to promote the migration, proliferation, and invasion abilities of lung cancer cells (9). LncRNA TUC338 has been shown to promote the invasion and progression of lung tumors by stimulating the mitogen-activated protein kinase pathway (10).
The lncRNA RUNX1-IT1 is a newly discovered non-coding RNA with important functions. The level of expression of lncRNA RUNX1-IT1 in endometrial cancer has been shown to be significantly reduced (11). Further, the expression of lncRNA RUNX1-IT1 can arrest the growth of liver cancer cells and induce their apoptosis (12). A low expression of lncRNA RUNX1-IT1 indicates a poor prognosis among liver cancer patients; thus, as a biomarker of liver cancer prognosis and a therapeutic target, lncRNA RUNX1-may be a powerful molecular target (10). Further, lncRNA RUNX1-IT1 has been shown to be down-regulated in colon cancer cell lines and tissues, and thus could be employed as a diagnostic biomarker and play a role in tumor inhibition (13). In lung tumor tissues and cell lines, the molecular mechanism, biological role, and expression level of lncRNA RUNX1-IT are still unknown. In this study, we sought to elaborate on the basic role and underlying mechanism of this gene in lung tumor tissues and in-vitro cell models. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-2210).
Methods
Sample collection and cell line
We gathered the data of 30 lung cancer patients who received treatment at the Second Affiliated Hospital of Nantong University’s Thoracic Surgery Department between May and September 2020. During surgical resection, tumor tissues and paired normal tissues adjoining the cancer (>5 cm away from the margin of the tumor mass) were obtained from patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Second Affiliated Hospital of Nantong University (No. 20201584) and informed consent was taken from all the patients.
All samples were washed with phosphate buffered saline immediately after sampling, and then placed in liquid nitrogen for the subsequent extraction of RNA. Human lung cancer cell lines (PC-9 cells, A549 cells, H460 cells, H1299 cells, and SPC-A1 cells) and a normal epithelial lung cell line (BEAS-2B) from a normal human were acquired from the Shanghai Cell Bank, Chinese Academy of Sciences. RPMI 1640 medium comprising 100 U/mL of penicillin, 10% fetal bovine serum (FBS), and 100 µg/mL of streptomycin was used for the culture at 37 °C and 5% carbon dioxide (CO2).
The expression of lncRNA RUNX1-IT1 was detected by quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from clinical samples of the tissues, and cells were cultivated using an RNA extraction kit in accordance with the manufacturer’s instructions. After extraction, the total RNA concentration was determined by NanoDrop 2000, and the expression levels of the target genes were quantitatively analyzed by Applied Biosystems 7500 Fast. The relative quantitative determination of each gene was performed using the double-standard curve approach. The 2−ΔΔCt approach was used to measure the relative expressions of each gene, where ΔCt = the target gene Ct value – the internal reference gene Ct value, and ΔΔCt = the transfection group ΔCt – the control group ΔCt, in which lncRNA RUNX1-IT1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primer sequences were used as references (14).
Vector construction and transfection
Small hairpin RNA (shRNA) targeting lncRNA RUNX1-IT1 was designed and synthesized for the negative control and the overexpression vector reference (12). Next, Lipofectamine 3000 was used for transfection, and a 6-well plate was inoculated with lung tumor cells that were in a state of good growth. After 12 h, when the cell density reached about 85–90%, the designed lncRNA RUNX1-IT1 interference sequences (shRNA-1 and shRNA-2) and negative control-shRNA were transfected into the cells with Lipofectamine 3000 transfection reagent. See the references for the sequences and overexpression sequences of the interfering primers (14). Transfection efficiency was detected by reverse transcription polymerase chain reaction (RT-PCR) after 48 h.
Cell activity was detected by Cell Counting Kit 8 (CCK-8)
Post-transfection, the lung tumor cells were harvested in the logarithmic growth phase, diluted into 3×104/mL cell suspension with Dulbecco’s Modified Eagle Medium comprising 10% FBS, and then inoculated into a 96-well culture plate with 100 µL per well. The cells were then incubated in a cell incubator at 37 °C and 5% CO2. After 72, 48, and 24 h of culture, 10 µL CCK-8 reagent was added. After 3 h of culture in a 37 °C incubator, the optical density (OD) value of each well at 490 nm was observed by a full wavelength microplate reader. The assessment was repeated 3 times. The absorbance values of the 5 complex pores were averaged, and a cell growth curve was constructed.
Clone creation assessment
For the inoculation procedure, the transfected cells were cultivated in 6-well plates, and the density of each well was 700 cells. The culture medium comprising 10% FBS was cultivated during the night, and the solution was changed every 3 days. The cells were fixed using methanol and stained with 0.1% crystal violet after 14 days. The number of clones was calculated after air drying.
Tumor cell suspension sphere-forming experiment
At approximately 48 h post-transfection, the lung tumor cells were collected and digested with 0.25% trypsin, centrifuged to collect cell precipitation, and then cultured completely with 10% FBS (containing 20 ng/mL epithelial growth factor, and 10 ng/mL fibroblast growth factor 2, N2 and B27) and blown into single-cell suspension. The cells were inoculated in a serum-free ultra-low attachment culture dish.
Transwell assay
The invasion ability of lung cancer cells was evaluated using a Transwell assay. First, the lung cancer cells were collected 48 h after transfection, digested with 0.25% trypsin, centrifuged to collect cell precipitation, and suspended with 1% complete medium. The cell density was then calculated. Next, 5×104 cells were inoculated into the upper chamber of the Transwell (BioCoat Matrigel) inserts in 24 wells. The lower chamber was incubated with a medium containing 20% serum for 24 h, the cells were then adjusted with 4% paraformaldehyde for 0.5 h, and subsequently stained with 0.5% crystal violet for 5 minutes. The number of cells invading the surface of the lower chamber was observed under an inverted microscope.
Western blot test
Transfected lung cancer cells were collected and lysed on ice after adding protease inhibitor, and the concentration of the proteins was assessed using the bicinchoninic acid assay protein quantitative method. The cells were transferred to polyvinylidene fluoride membranes by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, sealed in 5% skimmed milk powder in Tris buffer saline (pH 7.4) comprising 0.05% Tween 20 for 2 h at ambient temperature, and subsequently incubated with a primary antibody diluent at 4 °C for 12 h with GAPDH as an internal reference. Next, the cells were incubated with horseradish peroxidase (HRP) and a secondary antibody.
Statistical analysis
The statistical analysis was conducted using SPSS 20.0, a statistical computer program, and the results are presented as mean ± standard deviation (SD). Comparisons among the groups were conducted using a t-test or an analysis of variance. A P value <0.05 was considered statistically significant.
Results
Low expression of lncRNA RUNX1-IT1 in lung cancer tissues and cell lines
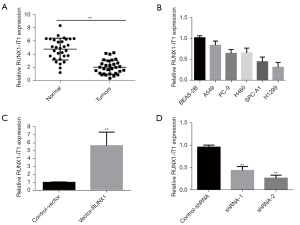
First, qRT-PCR was employed to examine the expression of lncRNA RUNX1-IT1 in 30 lung tumor tissues and neighboring tissues. The results showed that the lung tumor tissues had considerably lower levels of lncRNA RUNX1-IT1 expression than the neighboring tissues (P<0.01; see Figure 1A). Next, qRT-PCR was used to evaluate the expression levels of lncRNA RUNX1-IT1 in 5 lung tumor cell lines and in the normal lung epithelial cells (BEAS-2B) of a healthy human to further investigate the gene’s unique biological function in lung tumors. The results showed that the expression level of lncRNA RUNX1-IT1 was the highest in the BEAS-2B cells, and the expression levels of lncRNA RUNX1-IT1 were quite different in multiple lung tumor cell lines. Notably, the expression level was highest in the A549 cells and lowest in the H1299 cells (see Figure 1B). Thus, the overexpression vector of lncRNA RUNX1-IT1 was transfected in H1299 cells, and the interference vector of lncRNA RUNX1-IT1 was transfected in A549 cells. Expression efficiency was discerned by qRT-PCR. The results illustrated that the expression efficiency of lncRNA RUNX1-IT1 in H1299 cells in the lncRNA RUNX1-IT1 group was considerably superior to that of the Control-Vector group (see Figure 1C). Compared to the control group (control-shRNA), the expression efficiency of lncRNA RUNX1-IT1 in A549 cells decreased significantly after the transfection of the interference targets (shRNA-1 and shRNA-2) (see Figure 1D).
The effect of the knockdown or overexpression of lncRNA RUNX1-IT1 on the proliferation of lung cancer cells
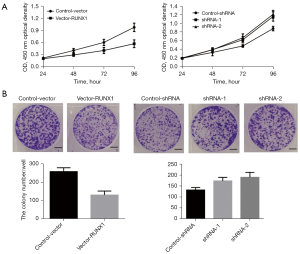
In relation to the role of lncRNA RUNX1-IT1 in lung tumor cells, the CCK-8 results showed that the overexpression of lncRNA RUNX1-IT1 in H1299 cells greatly limited the proliferation ability of lung tumor cells in comparison to cells in the control-shRNA group. Compared to the control-vector cells, shRNA-1 and shRNA-2 significantly promoted the growth of lung cancer cells in A549 cells at 96 h (see Figure 2A). The results of the clone formation experiment showed that in comparison to the control-vector cells, the overexpression of lncRNA RUNX1-IT1 in H1299 cells significantly inhibited the clone formation ability of lung cancer cells. Compared to cells in the control-shRNA group, shRNA-1 and shRNA-2 significantly promoted the number of lung cancer cell clones in A549 cells (see Figure 2B). As the above results indicate, lncRNA RUNX1-IT1, as a tumor suppressor gene, significantly regulated the proliferation ability of lung cancer.
The effect of overexpression or knockdown of lncRNA RUNX1-IT1 on the stemness of lung tumor cells
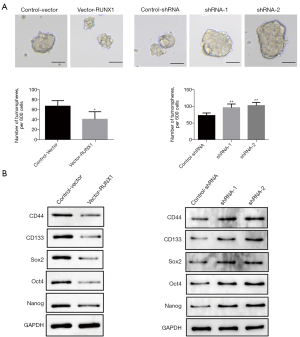
The above experiments showed that the overexpression or knockdown of lncRNA RUNX1-IT1 significantly affected the progression ability of lung tumor cells. To determine whether lncRNA RUNX1-IT1 affected the stemness of lung cancer cells, we examined the suspension of sphere forming in tumor cells, and found that in comparison to the control-vector cells, the overexpression of lncRNA RUNX1-IT1 in H1299 cells arrested the ability of lung tumor cells to suspend and spheroidize. In A549 cells, the knockdown of lncRNA RUNX1-IT1 (shRNA-1 and shRNA-2) promoted the ability of lung cancer cells to suspend and spheroidize (see Figure 3A). To further explore the molecular strategy of the effect of overexpression or knockdown of lncRNA RUNX1-IT1 on the stemness of lung cancer cells, we conducted a Western blot and found that compared to the control-vector cells, the overexpression of lncRNA RUNX1-IT1 in H1299 cells significantly inhibited the expressions of cluster of differentiation 44 (CD44), cluster of differentiation 133 (CD133), sex-determining region Y-box 2 (Sox2), octamer-binding transcription factor 4 (Oct4), and Nanog in lung cancer cells. Further, compared to the control-shRNA cells, the knockdown of lncRNA RUNX1-IT1 (shRNA-2 and shRNA-1) in A549 cells significantly promoted the expression of CD44, CD133, Sox2, Oct4, and Nanog in lung cancer cells (see Figure 3B).
The effect of the knockdown or overexpression of lncRNA RUNX1-IT1 on the invasion of lung tumor cells
The ability to invade and migrate is a main characteristic of tumor cells. Thus, a Transwell assay was used to further explore the effect of lncRNA NEAT1 on the invasion of lung tumor cells. Compared to the control-vector cells, the overexpression of lncRNA RUNX1-IT1 in H1299 cells significantly inhibited the invasion ability of lung cancer cells. Compared to the control-shRNA cells, the knockdown lncRNA RUNX1-IT1 (shRNA-1 and shRNA-2) in A549 cells significantly promoted the invasion ability of lung tumor cells (see Figure 4A). The effect of the knockdown or overexpression of lncRNA RUNX1-IT1 on the expression of key protein molecules of lung cancer cell invasion was evaluated by Western blot. Compared to the control-vector cells, the overexpression of lncRNA RUNX1-IT1 in H1299 cells significantly improved E-cadherin expression and arrested the expression of N-cadherin and vimentin. Compared to the control-shRNA cells, the knockdown of lncRNA RUNX1-IT1(shRNA-1 and shRNA-2) in A549 cells, significantly suppressed the expression level of E-cadherin, but significantly increased the expression levels of vimentin and N-cadherin (see Figure 4B).

Discussion
Recently published articles have shown that lncRNAs are strongly linked to tumor metastasis and invasion, and may be biomarkers for early tumor detection and treatment (15,16). For example, lncRNA H H19 is highly expressed in the cell lines of lung tumors and tissues, and lncRNA H19 overexpression significantly improves lung cancer proliferation and metastasis (17). Additionally, the high and low expressions of lncRNA SLC16A1-AS1 are significantly correlated with the overall survival time and the progression-free survival time of lung cancer patients, and lncRNA SLC16A1-AS1 a promising biomarker for lung cancer patients (18). LncRNA XLOC_009167 can be used as a new biomarker for the early detection of lung cancer, benign lung diseases, and healthy controls (19). Thus, clarifying the roles and functions of new lncRNAs in lung cancer will make lncRNAs biomarkers for targeted therapies, surgical monitoring, and the early detection of lung cancer. In the present study, we found that the low expression and overexpression or downregulation of lncRNA RUNX1-IT1 significantly arrested clone formation, proliferation, invasion, and the cell stemness of lung cancer cells in lung cancer tissues and cell lines. A further study showed that the overexpression or low expression of lncRNA RUNX1-IT1 significantly affected the expression of cell stemness-associated proteins and invasion-related proteins.
LncRNA RUNX1-IT1 is a newly identified non-coding RNA with an important regulatory function, and can inhibit or promote cancer in different tumors; for example, it is lowly expressed in liver cancer, endometrial cancer, and colorectal cancer, but it is highly expressed in pancreatic cancer and glioma (11,13,14,20,21). The lncRNA RUNX1-IT1 expression level in liver cancer tissues is significantly reduced, and the overexpression of lncRNA RUNX1-IT1 can limit the growth of liver cancer cells and induce their apoptosis. The low expression of lncRNA RUNX1-IT1 indicates a poor prognosis for liver cancer patients (20). Conversely, lncRNA RUNX1-IT1 is considerably upregulated in samples of pancreatic cancer patients, and is related to poor prognosis (14). The overexpression of RUNX1-IT1 has been found to promote the migration, proliferation, and invasion of pancreatic cancer and can be employed as a potential prognostic biomarker and treatment option for pancreatic carcinoma (14).
The results indicate that lncRNA RUNX1-IT1 may have different physiological functions in different tumors. Our previous study (data not published) showed that a total of 1,523 genes were differently regulated in primary lung tumors compared with normal tissue. We identified 201 genes that were subsequently up-regulated and 437 genes (including: lncRNA RUNX1-IT1) down-regulated during tumor progression.
In this study, we discovered that lncRNA RUNX1-IT1 was weakly expressed in lung tumor cell lines and tissues. The results showed that the expression level of LncRNA RUNX1-IT1 was highest in the A549 cells and lowest in the H1299 cells (Figure 1B). Therefore, the overexpression vector of lncRNA RUNX1-IT1 was transfected in H1299 cells, and the interference vector of lncRNA RUNX1-IT1 was transfected in A549 cells to study the role of lncRNA RUNX1-IT1(down-regulated or up-regulated) in lung tumor cells. The overexpression of lncRNA RUNX1-IT1 significantly inhibited the proliferation of cells and clone creation ability, while the knockdown of lncRNA RUNX1-IT1 significantly promoted the proliferation of cells and clone creation ability. Our findings revealed that lncRNA RUNX1-IT1 has a promising role as a tumor repressor gene in lung cancer.
Cancer stem cells (CSCs) with self-renewal and differentiation ability have been identified in multiple solid tumors, including glioma, lung cancer, and liver cancer (22-24). Recently published articles have stated that the CSCs of lung cancer have high tumorigenicity and self-renewal and differentiation ability, which are not only related to tumor metastasis and epithelial-mesenchymal transition, but to drug resistance and the recurrence of tumors (25,26). Thus, inhibiting tumor stem cells is a novel treatment strategy that could improve the survival time of patients. For example, lncRNA HAND2-AS1 inhibits migration, invasion and cell stemness of non-small cell lung cancer by interacting with transforming growth factor beta 1 (27). However, long-chain ncRNA Linc00662 is capable of promoting the invasion of lung cancer cells and maintaining the characteristics of tumor stem cells (28). Additionally, CD44 and CD133 are stem cells markers and are highly expressed in lung CSCs (22). Oct4, Sox2, and Nanog have been recognized as stem cell regulatory factors that perform essential tasks in preserving the self-renewal, proliferation, and multi-directional differentiation potential of stem cells (29,30). In this study, the overexpression or knockdown of lncRNA RUNX1-IT1 was found to regulate the stemness characteristics of lung cancer. The Western blot results showed that the overexpression of lncRNA RUNX1-IT1 arrested the expression of CD44, CD133, Sox2, Oct4, and Nanog. Additionally, the overexpression of lncRNA RUNX1-IT1 may also prevent the invasion of lung cancer cells by promoting the expression of E-cadherin. The above findings indicate that lncRNA RUNX1-IT1 plays an essential role in the maintenance and invasion of lung CSCs. Our present results showed overexpression or knockdown of lncRNA RUNX1-IT1 affects the stemness and invasion ability of lung cancer cells. It is well known that CSCs are a subpopulation of cancer cells that play a pivotal role in tumor development, invasion, metastasis, and recurrence (22-23). Therefore, activated-lncRNA RUNX1-IT1 compounds or agents might serve as novel therapeutic strategies for lung cancer treatment. In addition, qRT-PCR was employed to examine the expression of lncRNA RUNX1-IT1 in lung tumor tissues and evaluate the stemness and invasiveness of lung cancer.
However, the biological role of lncRNA RUNX1-IT1 in lung cancer is unclear. Thus, in future research, we will continue to explore whether lncRNA RUNX1-IT1 controls downstream target gene expression by intermediate miRNAs or other gene proteins.
Acknowledgments
Funding: The study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-2210
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-2210
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-2210). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Second Affiliated Hospital of Nantong University (No. 20201584) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schabath MB, Cote ML. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol Biomarkers Prev 2019;28:1563-79. [Crossref] [PubMed]
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. [Crossref] [PubMed]
- Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat Res 2016;170:25-46. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Gasparri R, Romano R, Sedda G, et al. Diagnostic biomarkers for lung cancer prevention. J Breath Res 2018;12:027111. [Crossref] [PubMed]
- Zito Marino F, Bianco R, Accardo M, et al. Molecular heterogeneity in lung cancer: from mechanisms of origin to clinical implications. Int J Med Sci 2019;16:981-9. [Crossref] [PubMed]
- Hu J, Wang T, Chen Q. Competitive endogenous RNA network identifies four long non-coding RNA signature as a candidate prognostic biomarker for lung adenocarcinoma. Transl Cancer Res 2019;8:1046-64. [Crossref]
- Jathar S, Kumar V, Srivastava J, et al. Technological Developments in lncRNA Biology. Adv Exp Med Biol 2017;1008:283-323. [Crossref] [PubMed]
- Li J, Xia R, Liu T, et al. LncRNA-ATB Promotes Lung Squamous Carcinoma Cell Proliferation, Migration, and Invasion by Targeting microRNA-590-5p/NF90 Axis. DNA Cell Biol 2020;39:459-73. [Crossref] [PubMed]
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci 2018;22:443-9. [PubMed]
- Liang M, Wang H, Liu C, et al. LncRNA RUNX1-IT1 is Downregulated in Endometrial Cancer and Binds to miR-21 Precursor to Suppress Its Maturation. Cancer Manag Res 2020;12:13451-9. [Crossref] [PubMed]
- Sun L, Wang L, Chen T, et al. LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. Cell Death Dis 2020;11:95. [Crossref] [PubMed]
- Shi J, Zhong X, Song Y, et al. Long non-coding RNA RUNX1-IT1 plays a tumour-suppressive role in colorectal cancer by inhibiting cell proliferation and migration. Cell Biochem Funct 2019;37:11-20. [Crossref] [PubMed]
- Liu S, Zhang J, Yin L, et al. The lncRNA RUNX1-IT1 regulates C-FOS transcription by interacting with RUNX1 in the process of pancreatic cancer proliferation, migration and invasion. Cell Death Dis 2020;11:412. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Dastmalchi N, Safaralizadeh R, Nargesi MM. LncRNAs: Potential Novel Prognostic and Diagnostic Biomarkers in Colorectal Cancer. Curr Med Chem 2020;27:5067-77. [Crossref] [PubMed]
- Zhao Y, Feng C, Li Y, et al. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem 2019;460:1-8. [Crossref] [PubMed]
- Liu HY, Lu SR, Guo ZH, et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J Investig Med 2020;68:52-9. [Crossref] [PubMed]
- Jiang N, Meng X, Mi H, et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin Chim Acta 2018;486:26-33. [Crossref] [PubMed]
- Yan PH, Wang L, Chen H, et al. LncRNA RUNX1-IT1 inhibits proliferation and promotes apoptosis of hepatocellular carcinoma by regulating MAPK pathways. Eur Rev Med Pharmacol Sci 2019;23:8287-94. [PubMed]
- Wu Z. MiR-195 connects lncRNA RUNX1-IT1 and cyclin D1 to regulate the proliferation of glioblastoma cells. Int J Neurosci 2021; [Epub ahead of print]. [Crossref] [PubMed]
- Prabavathy D, Ramadoss N. Heterogeneity of Small Cell Lung Cancer Stem Cells. Adv Exp Med Biol 2019;1139:41-57. [Crossref] [PubMed]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, et al. Cancer stem cells in glioblastoma. Genes Dev 2015;29:1203-17. [Crossref] [PubMed]
- Liu YC, Yeh CT, Lin KH. Cancer Stem Cell Functions in Hepatocellular Carcinoma and Comprehensive Therapeutic Strategies. Cells 2020;9:1331. [Crossref] [PubMed]
- Leon G, MacDonagh L, Finn SP, et al. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther 2016;158:71-90. [Crossref] [PubMed]
- Ren W, Hou J, Yang C, et al. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J Exp Clin Cancer Res 2019;38:62. [Crossref] [PubMed]
- Miao F, Chen J, Shi M, et al. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-β1. Biosci Rep 2019;39:BSR20181525. [Crossref] [PubMed]
- Gong W, Su Y, Liu Y, et al. Long non-coding RNA Linc00662 promotes cell invasion and contributes to cancer stem cell-like phenotypes in lung cancer cells. J Biochem 2018;164:461-9. [Crossref] [PubMed]
- Keshavarz M, Asadi MH. Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J 2019;286:2611-23. [Crossref] [PubMed]
- Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep 2017;50:117-25. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


