
Exploring the molecular targets and mechanisms of [10]-Gingerol for treating triple-negative breast cancer using bioinformatics approaches, molecular docking, and in vivo experiments
Introduction
Breast cancer is the most frequent cancer in women worldwide and has caused the highest cancer deaths in 2020 (1). In developing countries, it has the highest morbidity and mortality in women (2). Triple-negative breast cancer (TNBC) accounts for 20–30% of breast cancer cases and is characterized by the lack of estrogen (ER), progesterone (PR) and human epidermal growth factor receptor-2 (HER2) receptor expression, and its rapid progression to metastasis (2-4). Due to the lack of receptors’ expression for estrogen, progesterone, or HER2, the patients with TNBC do not respond to endocrine or HER2-targeted therapies, leaving chemotherapy as the only option for the primary treatment of TNBC. TNBC is initially very sensitive to chemotherapy; relapse is common with the emergence of chemotherapy-resistant metastases, inevitably leading to death (5-7). Therefore, the long-term efficacy of chemotherapy for advanced TNBC is limited and the TNBC patients and their normal tissue toxicity remain a major concern. Therefore, the search for natural drugs with fewer side effects is needed as an alternative or complementary chemotherapy for advanced breast cancer.
Ginger (Zingiber officinale Roscoe, Zingiberaceae) is a natural dietary rhizome of the plant and is widely used as a traditional herb and flavoring agent (8). Ginger contains a variety of biologically active ingredients, such as gingerol, shogaols, polyphenols, and gingerone, indicating its role in mediating the anti-inflammatory and anti-tumor activities (9-12). Among the bioactive components of ginger, [6]-Gingerol and [8]-Gingerol have been widely reported to exhibit antitumor activity in multiple tumors by inhibiting the proliferation and migration of cells and inducing the apoptosis of tumor cells (13-16). [10]-Gingerol is one of the main phenolic compounds isolated from ginger and can inhibit the proliferation of tumor cells and induce the apoptosis of ovarian, colon, and prostate cancer cells (17-19). Although [10]-Gingerol has been reported to exhibit anti-breast cancer cell activity, its molecular mechanism is poorly understood (20). In this study, first, apoptosis and proliferation assays were used to identify the anti-cancer ability and pro-apoptotic pathways for [10]-Gingerol in TNBC. Then, the network pharmacology and molecular biology approaches were used to investigate the bioactive compounds and underlying mechanisms of ginger and [10]-Gingerol for the treatment of TNBC. This study demonstrated potential mechanisms that might underlie the therapeutic effects of ginger’s potent constituent, [10]-Gingerol, against TNBC and provided evidence to support its clinical use. We present the following article in accordance with the MDAR reporting checklist (Available at https://dx.doi.org/10.21037/tcr-21-1138).
Methods
Identification of [10]-Gingerol as the key component of ginger
The bioactive compounds of ginger were obtained from the Traditional Chinese Medicine System Pharmacology database and analysis platform (TCMSP, http://lsp.nwu.edu.cn/). The main bioactive compounds of ginger were screened using criteria OB ≥15% and DL ≥0.15, among which [10]-Gingerol was one of the important components of ginger (Table 1).
Table 1
| Target name | Compound name | BC | CC | DG | OB, % | DL |
|---|---|---|---|---|---|---|
| MOL002459 | 10-gingerol | 0.029 | 0.398 | 20 | 19.14 | 0.28 |
| MOL000449 | Stigmasterol | 0.029 | 0.482 | 11 | 43.83 | 0.76 |
| MOL000358 | beta-sitosterol | 0.002 | 0.385 | 8 | 36.91 | 0.75 |
| MOL002467 | 6-gingerol | 0.002 | 0.376 | 5 | 35.64 | 0.16 |
| MOL003358 | Euxanthone | 0.009 | 0.418 | 5 | 92.98 | 0.16 |
BC, betweenness centrality; CC, closeness centrality; DG, degree; OB, oral bioavailability; DL: drug-likeness.
Therapeutic targets for ginger
In order to obtain the putative targets of ginger, PubChem (https://pubchem.ncbi.nlm.nih.gov/) and TCMSP databases were used. The canonical SMILES strings for all the chemical compounds of ginger were obtained from PubChem. These canonical SMILES were submitted into the TCMSP database to obtain the main targets of ginger. The duplications in compounds were deleted to obtain the final target of ginger.
Potential target genes of TNBC
The mRNA expression data of TNBC and adjacent tissues were downloaded from the Cancer Genome Atlas (TCGA) website (https://portal.gdc.cancer.) and the datasets GSE76124 and GSE112825 were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/). Then, the Omicshare (https://www.omicshare.com/tools/index.php/) online system was used to normalize the included datasets.
R ‘limma’ package (version 3.32.10) (21) was used to assess differentially expressed genes (DEGs). DEGs were screened according to P<0.05 and |log2 FC| >1.0. The screening of DEGs resulted in obtaining the TNBC-associated genes.
Protein-protein interaction (PPI) network
The DEGs, obtained from GEO and TCGA databases, and ginger targets were imported to Venny (https://bioinfogp.cnb.csic.es/tools/venny/) online software to obtain the intersection of three datasets. The overlapped genes were potential targets for the ginger intervention in TNBC. In order to obtain a PPI network for the potential targets of ginger intervention for TNBC, these overlapped targets were imported into STRING (https://string-db.org/) database; the human was set as target species and the confidence score of 0.400 was selected as a threshold value. Finally, Cytoscape software version 3.8.1 (22) was used to visualize the PPI network.
Construction of components-targets network and screening of core targets
The corresponding targets of ginger’s bioactive compounds and the PPI network of potential targets of ginger intervention for TNBC were imported into Cytoscape software version 3.8.1 (22) to build a “components-targets” network. Then, the “Network Analyzer”, a plug-in that comes with Cytoscape, was used to analyze the topology properties of the “components-targets” network. The core targets were selected with the node >2-fold Degree (DG), >1-fold Betweenness Centrality (BC), and >1-fold Closeness Centrality (CC).
GO and KEGG pathway enrichment analysis
DAVID (https://david.ncifcrf.gov/) (23) was used to conduct the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the related coding genes. The GO analyses included biological process (BP), cellular component (CC), and molecular function (MF). Significant pathways and biological functions were screened according to P<0.05.
Survival analysis
The data obtained from the TCGA dataset were used to validate the survival effects. In this study, R ‘survminer’ package (https://CRAN.R-project.org/package=survminer) was used to perform the overall survival analysis of the above core targets.
Molecular docking
The 3D structures of the core targets obtained in the previous step were downloaded from the Protein Data Bank database (PDB; https://www.rcsb.org/) and the MOL2 structure file for each key compound was obtained from the TCMSP database. Ligand-target separation was performed on each target using Discovery Studio software for obtaining the protein and ligand structures of each target. The proteins were prepared for docking by dehydrating, hydrogenating, calculating the charge, and assigning atomic type. The ligands were prepared by hydrogenating, adding charge, removing non-polar hydrogen, deleting lone pair electrons, and establishing special rotatable bonds. Then, AutoDock software was used to set binding pockets for molecular docking using the original ligands of each target as a reference. Finally, the AutoDock Vina was used to conduct the molecular docking of each target protein with ligand and the binding energy data were obtained for analysis.
Cell culture and the preparation of [10]-Gingerol
Human TNBC cell lines MDA‐MB‐231 and non-tumor breast cell line MCF-10A were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM), which was supplemented with 10% fetal bovine serum (FBS), and incubated in an incubator (WCI-180, Wiggens, Germany) with a humid atmosphere at 37 °C and 5% CO2. All the cell culture reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA). [10]-Gingerol was purchased from Chengdu Ruifensi Biotechnology Co., Ltd. (Chengdu, China).
Cell proliferation assay
The viability of [10]-Gingerol in TNBC cells was analyzed using a CCK‐8 assay following the manufacturer’s instructions. The MDA‐MB‐231 cells were inoculated into 96-well plates with a cell density of 4×103 per well for 24 hours. After 24 hours, the different concentrations of [10]-Gingerol compounds (0, 50, 75, 100, 125, and 150 µmol/L) were used to treat the cells with 3 holes in each group. After 24 hours of incubation, 10 μL of CCK-8 solution was added to the cells for 2 hours at 37 °C. The OD value was measured using the enzyme labeling instrument at 450 nm. Each experiment was carried out in triplicate; the half inhibitory concentration (IC50) was calculated using Prism 7 (GraphPad software).
Cell apoptosis assay by flow cytometry
The breast cancer cells, at the logarithmic growth phase, were inoculated on a 6-well plate at the density of 1×105 cells per well; after adhering to the wall, the cells were treated with the indicated doses of [10]-Gingerol. After 48 hours, the cells were collected. According to Annexin-V-FITC/PI staining instructions (KeyGEN Biotech, Jiangsu, China), 5-μL FITC Annexin V and 5-μL PI were added to each tube and incubated in dark for 15 min. Then, 200-μL Binding Buffer was added. Apoptosis was detected using Cytomic FC500 flow cytometry (Beckman Coulter, Inc., Brea, CA). The data were analyzed using NovoExpress software.
Western blot (WB)
After flow cytometry analysis, the cells were collected for lysis. Protein concentration was determined using Keygen BCA Protein Assay Kit (KEYGEN, China). An equivalent amount of 20 μg protein was disintegrated using 10% SDS-PAGE and then transferred to the PVDF membrane. The membrane was blocked with 5% blocking buffer (5% skimmed milk powder + TBST) for 60 min at room temperature. Subsequently, the membrane was incubated with primary antibodies, including Caspase-3 antibody (Cell Signaling Technology, CST#9662S; 1:1,000), Caspase-7 antibody (CST#12827S; 1:1,000), and Caspase-8 antibody (CST#4790S; 1:1,000), overnight at 4 °C. The secondary antibodies were then added on the second day and incubated for 60 min. After the membranes were washed, the bands were visualized using ChemiDoc TMXRS+ (Bio-RAD, USA). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
All the measurements were taken in triplicates and the data were expressed as mean ± The Student’s t‐test was used to compare the statistical differences the between two groups. The comparison of multiple groups was carried out using analysis of variance (ANOVA) followed by the Tukey range test to analyze the differences between conditions. In each case, the value of P<0.05 was considered statistically significant.
Results
Identification of potential targets of ginger and TNBC
After differential analysis, a total of 3,228 DEGs (1,250 up-regulated and 1,977 down-regulated) were identified in the TCGA database, while 3,674 DEGs (2,745 up-regulated and 929 down-regulated) were identified in the GEO database and are presented as volcano plots (Figure 1A,1B). The “up-regulated” or “down-regulated” polarity of genes in this article is described as tumor vs. normal.
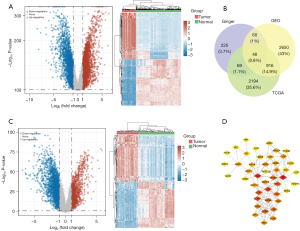
A total of 265 compounds of ginger were obtained from the TCMSP database (Available online: https://cdn.amegroups.cn/static/public/tcr-21-1138-1.xlsx). According to the results of PubChem and TCMSP databases, a total of 402 therapeutic targets were identified for all the compounds of ginger. Meanwhile, Venn Diagram online software was used to determine the intersection of DEGs from TCGA, GEO, and ginger targets (Figure 1C). A total of 48 genes were identified as potential targets in the three data sets, which suggested that ginger had 48 therapeutic targets for TNBC (Figure 1D). Moreover, these 48 therapeutic targets corresponded to 117 compounds of ginger (Available online: https://cdn.amegroups.cn/static/public/tcr-21-1138-2.xlsx).
PPI network analysis and screening of core targets
A total of 108 potential targets were submitted to the STRING database to build a PPI network (Figure 2A). The network was analyzed with Network Analyzer. The core targets were selected with the node >2-fold DG, >1-fold BC, and >1-fold CC. Among them, a total of 29 therapeutic targets were identified, including ESR1, ADH1C, MAOB, ADRB2, PPARG, NR3C1, ADH1B, CCNA2, PGR, IL6, ADRB1, JUN, CAT, MAOA, CHEK1, ADRA2A, CCNB1, CDK1, AURKA, LPL, MAPK10, LIPE, CDK5, SHMT2, ALDH1A1, NR3C2, GPI, FASN, and PTGER3, which indicated that these targets might be the core therapeutic targets for TNBC by ginger (Table 2, Figure 2B).
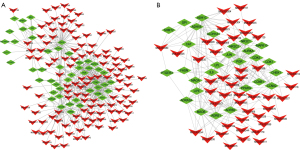
Table 2
| Target name | Betweenness centrality | Closeness centrality | Degree |
|---|---|---|---|
| ESR1 | 0.370 | 0.526 | 74 |
| ADH1C | 0.234 | 0.473 | 45 |
| MAOB | 0.176 | 0.449 | 34 |
| ADRB2 | 0.070 | 0.425 | 31 |
| PPARG | 0.061 | 0.442 | 30 |
| NR3C1 | 0.072 | 0.457 | 30 |
| ADH1B | 0.082 | 0.451 | 29 |
| CCNA2 | 0.014 | 0.415 | 23 |
| PGR | 0.016 | 0.446 | 22 |
| IL6 | 0.055 | 0.458 | 20 |
| ADRB1 | 0.021 | 0.384 | 18 |
| JUN | 0.023 | 0.448 | 18 |
| CAT | 0.124 | 0.513 | 17 |
| MAOA | 0.039 | 0.454 | 16 |
| CHEK1 | 0.017 | 0.385 | 16 |
| ADRA2A | 0.023 | 0.389 | 15 |
| CCNB1 | 0.011 | 0.430 | 13 |
| CDK1 | 0.003 | 0.374 | 12 |
| AURKA | 0.006 | 0.404 | 11 |
| LPL | 0.007 | 0.402 | 10 |
| MAPK10 | 0.006 | 0.433 | 9 |
| LIPE | 0.004 | 0.366 | 9 |
| CDK5 | 0.001 | 0.355 | 9 |
| SHMT2 | 0.009 | 0.356 | 7 |
| ALDH1A1 | 0.033 | 0.453 | 7 |
| NR3C2 | 0.000 | 0.369 | 6 |
| GPI | 0.007 | 0.355 | 6 |
| FASN | 0.013 | 0.378 | 6 |
| PTGER3 | 0.006 | 0.374 | 4 |
Functional enrichment analysis of core targets
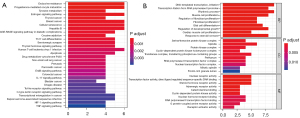
KEGG and GO analyses were performed for all the 29 core targets. For KEGG pathway analysis, the hub genes were mainly enriched in endocrine resistance, progesterone-mediated oocyte maturation, estrogen signaling pathway, breast cancer, and cellular senescence (Figure 3A). GO analysis included biological process (BP), cellular component (CC), and molecular function (MF). The top five BP terms for GO analysis included a response to DNA-templated transcription, initiation, rhythmic process, transcription initiation from RNA polymerase II promoter, muscle cell proliferation, and regulation of developmental growth (Figure 3B). The top five CC terms for GO analysis included spindle, transferase complex, transferring phosphorus-containing groups, serine/threonine-protein kinase complex, protein kinase complex, and perikaryon. The top five MF terms for GO analysis included nuclear receptor activity, transcription factor activity, direct ligand regulated sequence-specific DNA binding, steroid hormone receptor activity, nuclear hormone receptor binding, and RNA polymerase II transcription factor binding.
Overall survival analysis of core targets
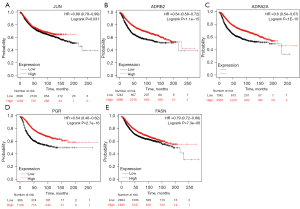
In order to assess whether the mRNA levels of core targets harbored prognostic implications for TNBC, the RNA-seq data of TNBC patients obtained from the TCGA database was analyzed and found that the low expressions of JUN (HR =0.88, log-rank P=0.031), ADRB2 (HR =0.64, log-rank P<0.001), ADRA2A (HR =0.6, log-rank P<0.001), PGR (HR =0.54, log-rank P<0.001), and FASN (HR =0.79, log-rank P<0.001) were significantly associated with lower overall survival (Figure 4).
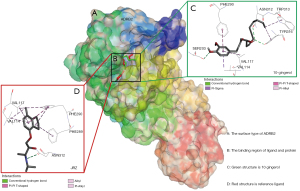
Docking results of [10]-Gingerol hub genes
The [10]-Gingerol was docked with 5 hub targets that harbored prognostic implications, including ADRA2A, ADRB2, PGR, JUN, and ALDOA. The binding energies are listed in Table 3. As compared to the reference ligand, the binding energies of [10]-Gingerol with ADRB2, ADRA2A, and ALDOA were generally ideal. Of these, the affinity by ADRB2 performed best and the docking patterns of [10]-Gingerol with ADRB2 are presented in Figure 5.
Table 3
| Ligands targets | Affinity (kcal/mol) | ||||
|---|---|---|---|---|---|
| ADRA2A (6k42) | ADRB2 (3ny8) | FASN (2cg5) | JUN (4hmy) | PGR (1a28) | |
| Reference ligands | |||||
| CZX | −7 | ||||
| JRZ | −7.9 | ||||
| COA | −6.3 | ||||
| GTP | −6.6 | ||||
| STR | −11 | ||||
| [10]-Gingerol | −7.1 | −8 | −7.8 | −6 | −7 |
[10]-Gingerol effectively inhibits the growth of TNBC cells
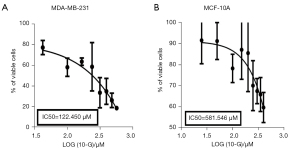
The effect of [10]-Gingerol on the cellular viability of MDA-MB-231 and MCF-10A was detected using the CCK-8 assay. Cells were exposed to the various concentrations (50–150 μM) of [10]-Gingerol for 24 hours and their viabilities were determined. As shown in Figure 6, [10]-Gingerol significantly inhibited the proliferation of MDA-MB-231 cells in a dose-dependent manner. After 24 hours of exposure, the IC50 value of [10]-Gingerol was 122.450 μM (Figure 7A). In comparison, the lowest inhibition was observed in the [10]-Gingerol-treated MCF-10A cells, where the IC50 value was 581.546 μM (Figure 7B). Therefore, [10]-Gingerol exhibited strong inhibitory effects on the growth of TNBC cells, while had low inhibition on healthy breast epithelial cells.
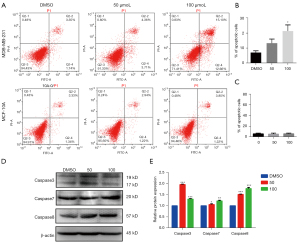
[10]-Gingerol specifically induces apoptosis in TNBC cells
The role of [10]-Gingerol in the induction of apoptosis in TNBC cells was analyzed. Flow cytometry analysis validated that the treatment of cells with [10]-Gingerol significantly increased the percentage of apoptotic cells as compared to the untreated control cells. The total apoptosis rates in the MDA-MB-231 human TNBC cells were 7.3%, 13.2%, and 21.9%, when the cells were treated with [10]-Gingerol at the concentrations of 0, 50, and 100 μM, respectively (Figure 6A,6B). The apoptosis rate in the MCF-10A cells was 6.5%, 6.3%, and 6.8%, when the cells were treated with [10]-Gingerol at the concentrations of 0, 50, and 100 μM, respectively (Figure 6A,6C). The results suggested that the apoptotic rate in TNBC cells induced by [10]-Gingerol increased with increasing the dose. In contrast, [10]-Gingerol had lesser pronounced pro-apoptotic effects on normal breast epithelial MCF-10A cells.
In order to study the apoptotic effects of [10]-Gingerol on TNBC cells, the expression levels of several apoptosis-related proteins, including Caspase-3, Caspase-7, and Caspase-8, were examined. The western blotting analysis demonstrated that, as compared to the DMSO group, after [10]-Gingerol treatment, the expression of Caspase-3, Caspase-7, and Caspase-8 increased (Figure 6D,6E). Notably, the up-regulatory effects of [10]-Gingerol on Caspase-7 and Caspase-8 increased with increasing the dose, suggesting a dose-dependent effect on their expression levels in TNBC cells. The result above indicated that [10]-Gingerol could inhibit the growth of TNBC by inducing apoptosis.
Discussion
The possible anticancer potential of ginger is receiving a great deal of attention. Information on the potential mechanism of ginger’s bioactive compounds, such as [10]-Gingerol, is important for understanding the biological effects of ginger. This study indicated that [10]-Gingerol could inhibit TNBC through the induction of apoptosis by activating the caspase-family protein. Moreover, combined with the network pharmacology and molecular docking analysis, this study demonstrated for the first time that [10]-Gingerol could inhibit the proliferation of TNBC cells and induce their apoptosis through the ADRB2 pathway.
Fresh ginger is rich in [6]-Gingerol, [8]-Gingerol, and [10]-Gingerol but most of the studies, reporting the anti-tumor ability of gingerols, are focused on [6]-Gingerol. However, some studies have illustrated that [10]-Gingerol has more significant anti-tumor activity than that of [6]-Gingerol (24). In this study, the anti-tumor activities of [10]-Gingerol were investigated against TNBC cells in vitro and demonstrated that [10]-Gingerol could inhibit the proliferation of MDA-MB-231 cells with an IC50 value of 122.450±0.5 μM. Moreover, this concentration had no obvious toxicity to non-tumor breast cell line (MCF-10A). In this study, multiple in vitro experiments were conducted on the effect of [10]-Ginger on TNBC cells and compared to the non-tumor MCF-10A breast cells. The IC50 value for non-tumor cytotoxicity was 581.546±2.5 μM, with the selectivity index (SI) of 4.78 (IC50 of non-tumor cells/IC50 of tumor cells), which is a high value as compared to the standard chemotherapeutics used in the clinic. These results confirmed that [10]-Gingerol had better tumor suppressor ability in TNBC.
Notably, recent studies have demonstrated that [10]-Gingerol can inhibit the growth of TNBC cells by affecting its cell cycle, but the results from these studies are contradictory. A study reported that [10]-Gingerol could inhibit the growth of TNBC cells at the concentration of 10 μM in vitro (25), while another study showed that [10]-Gingerol could inhibit the growth of TNBC cells at the concentration of >50 μM in vitro (26). Furthermore, another study reported that [10]-Gingerol could inhibit the growth of TNBC cells at the concentration of 666.2 μM in vitro (27). The IC50, which was obtained in this study, was different from the other studies, which might be due to differences in the production process of [10]-Gingerol in different companies, leading to different purities of [10]-Gingerol.
Moreover, [10]-Gingerol has been reported to exhibit anti-tumor activity by inducing apoptosis through the MAPK pathway in HCT116 human colon cancer cells (18). Gingerol has been subsequently shown to induce apoptosis in a variety of cancer cell types (17,28-30). In this study, [10]-Gingerol induced apoptosis by activating the caspase-family proteins (Caspase-3, Caspase-7, and Caspase-8).
Furthermore, the information from literature and publicly available databases were integrated to find the potential therapeutic protein targets for the TNBC treatment by ginger. A total of 265 bioactive compounds of ginger were found from previous studies, including [10]-Gingerol and others, among which 29 core targets of ginger for the treatment of TNBC were screened out. Enrichment analysis showed that endocrine resistance, progesterone-mediated oocyte maturation, estrogen signaling pathway, and cellular senescence were the potential target pathways for ginger’s bioactive compounds to inhibit the growth of TNBC cells.
Five pivotal proteins (ADRA2A, ADRB2, PGR, JUN, and FASN) not only had high BC and DG values in the PPI network but also identified as prognostic risk factors, suggesting that they might be the hub regulatory factors in the response of TNBC cells to ginger. In previous studies, these pivotal proteins have already been shown to participate in the pathophysiology and treatment of cancer. ADRA2A and ADRB2 belong to adrenoceptors and can regulate stress hormones. The high expression of ADRA2A and ADRB2 were related to the increased distant metastasis‐free survival (DMFS); ADRA2A was related to the inhibition of tumor cell proliferation, while ADRB2 was involved in anti-tumor immunity (31). However, studies have demonstrated that dexmedetomidine, an ADRA2A agonist, could promote metastasis in breast cancer via triggering the prolactin signaling pathway (32,33). Moreover, the blockage of ADRB2 was reported to improve the apoptosis in pancreatic cancer cells, suggesting that [10]-Gingerol could improve the TNBC apoptosis via regulating ADRB2 (34). PGR is a ligand-activated nuclear transcription factor that mediates progesterone activity; it is the most important prognostic and predictive immunoinflammatory marker in breast cancer (35). The variants of PGR were found to be associated with increased breast cancer susceptibility (36). FASN is involved in the de novo lipogenesis and regulation of ERα signaling (37). FASN is overexpressed in breast cancer cells and its silencing could result in the suppression of cell growth, but it also enhanced the cell invasion (38,39). JUN is an oncogenic transcription factor, which is an important regulator for a wide range of biological processes, such as the proliferation, differentiation, invasion, migration, and apoptosis of cells (40). JUN is highly expressed in various tumors, providing feedback on environmental stimuli, such as DNA damage (41).
This evidence supports that ginger might work against TNBC by regulating the expression of these hub genes. The current study aided in understanding the pathophysiology and roles of these genes in TNBC.
A molecular docking approach was also used to predict specific interactions between [10]-Gingerol and these five pivotal target proteins for ginger. The results showed that [10]-Gingerol could exert therapeutic effects against TNBC by targeting ADRB2, ADRA2A, and FASN. Among them, ADRB2 exhibited the highest affinity for [10]-Gingerol. The current results suggested that ginger and compound [10]-Gingerol might be the promising anti-cancer agent. Stress hormones or glycolysis might alternative effective approaches for these compounds to inhibit the growth of TNBC.
Conclusions
To sum up, [10]-Gingerol is a compound of ginger, which inhibited the proliferation of TNBC and induced apoptosis by activating the caspase-family protein. Moreover, the combination of network pharmacology method and molecular docking illuminated the molecular and pharmacological mechanisms of ginger and [10]-Gingerol against TNBC. ADRA2A, ADRB2, PGR, JUN, and FASN might play key roles in the effects of ginger against TNBC; among them, ADRA2A, ADRB2, and FASN might be the main targets for [10]-Gingerol in TNBC treatment. The current study might inspire and guide further work to verify the molecular targets of ginger and [10]-Gingerol against TNBC and its development in clinical use.
Acknowledgments
Funding: The study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1138
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-1138
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1138). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Ahmad A. Breast Cancer Statistics: Recent Trends. Adv Exp Med Biol 2019;1152:1-7. [Crossref] [PubMed]
- Valachis A, Nyström P, Fredriksson I, et al. Treatment patterns, risk for hospitalization and mortality in older patients with triple negative breast cancer. J Geriatr Oncol 2021;12:212-8. [Crossref] [PubMed]
- Bergin ART, Loi S. Triple-negative breast cancer: recent treatment advances. F1000Res 2019; [Crossref] [PubMed]
- Keenan TE, Tolaney SM. Role of Immunotherapy in Triple-Negative Breast Cancer. J Natl Compr Canc Netw 2020;18:479-89. [Crossref] [PubMed]
- Parsons HA, Burstein HJ. Adjuvant Capecitabine in Triple-Negative Breast Cancer: New Strategies for Tailoring Treatment Recommendations. JAMA 2021;325:36-8. [Crossref] [PubMed]
- Marquette C, Nabell L. Chemotherapy-resistant metastatic breast cancer. Curr Treat Options Oncol 2012;13:263-75. [Crossref] [PubMed]
- Banihani SA. Ginger and Testosterone. Biomolecules 2018;8:119. [Crossref] [PubMed]
- Ali BH, Blunden G, Tanira MO, et al. Some phytochemical, pharmacological and toxicological properties of ginger (Zingiber officinale Roscoe): a review of recent research. Food Chem Toxicol 2008;46:409-20. [Crossref] [PubMed]
- Prasad S, Tyagi AK. Ginger and its constituents: role in prevention and treatment of gastrointestinal cancer. Gastroenterol Res Pract 2015;2015:142979. [Crossref] [PubMed]
- Semwal RB, Semwal DK, Combrinck S, et al. Gingerols and shogaols: Important nutraceutical principles from ginger. Phytochemistry 2015;117:554-68. [Crossref] [PubMed]
- Mahomoodally MF, Aumeeruddy MZ, Rengasamy KRR, et al. Ginger and its active compounds in cancer therapy: From folk uses to nano-therapeutic applications. Semin Cancer Biol 2021;69:140-9. [Crossref] [PubMed]
- Xu S, Zhang H, Liu T, et al. 6-Gingerol suppresses tumor cell metastasis by increasing YAPser127 phosphorylation in renal cell carcinoma. J Biochem Mol Toxicol 2021;35:e22609. [Crossref] [PubMed]
- Hu SM, Yao XH, Hao YH, et al. 8-Gingerol regulates colorectal cancer cell proliferation and migration through the EGFR/STAT/ERK pathway. Int J Oncol 2020;56:390-7. [PubMed]
- Ma RH, Ni ZJ, Zhu YY, et al. A recent update on the multifaceted health benefits associated with ginger and its bioactive components. Food Funct 2021;12:519-42. [Crossref] [PubMed]
- Akamine LA, Vargas Medina DA, Lanças FM. Magnetic solid-phase extraction of gingerols in ginger containing products. Talanta 2021;222:121683. [Crossref] [PubMed]
- Rasmussen A, Murphy K, Hoskin DW. 10-Gingerol Inhibits Ovarian Cancer Cell Growth by Inducing G2 Arrest. Adv Pharm Bull 2019;9:685-9. [Crossref] [PubMed]
- Ryu MJ, Chung HS. 10-Gingerol induces mitochondrial apoptosis through activation of MAPK pathway in HCT116 human colon cancer cells. In Vitro Cell Dev Biol Anim 2015;51:92-101. [Crossref] [PubMed]
- Liu CM, Kao CL, Tseng YT, et al. Ginger Phytochemicals Inhibit Cell Growth and Modulate Drug Resistance Factors in Docetaxel Resistant Prostate Cancer Cell. Molecules 2017;22:1477. [Crossref] [PubMed]
- Fuzer AM, Martin A, Becceneri AB, et al. [10]-Gingerol Affects Multiple Metastatic Processes and Induces Apoptosis in MDAMB- 231 Breast Tumor Cells. Anticancer Agents Med Chem 2019;19:645-54. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Zhang F, Zhang JG, Qu J, et al. Assessment of anti-cancerous potential of 6-gingerol (Tongling White Ginger) and its synergy with drugs on human cervical adenocarcinoma cells. Food Chem Toxicol 2017;109:910-22. [Crossref] [PubMed]
- Joo JH, Hong SS, Cho YR, et al. 10-Gingerol inhibits proliferation and invasion of MDA-MB-231 breast cancer cells through suppression of Akt and p38MAPK activity. Oncol Rep 2016;35:779-84. [Crossref] [PubMed]
- Bernard MM, McConnery JR, Hoskin DW. [10]-Gingerol, a major phenolic constituent of ginger root, induces cell cycle arrest and apoptosis in triple-negative breast cancer cells. Exp Mol Pathol 2017;102:370-6. [Crossref] [PubMed]
- Martin AC, Fuzer AM, Becceneri AB, et al. [10]-gingerol induces apoptosis and inhibits metastatic dissemination of triple negative breast cancer in vivo. Oncotarget 2017;8:72260-71. [Crossref] [PubMed]
- Kim MO, Lee MH, Oi N, et al. 6-shogaol inhibits growth and induces apoptosis of non-small cell lung cancer cells by directly regulating Akt1/2. Carcinogenesis 2014;35:683-91. [Crossref] [PubMed]
- Ediriweera MK, Moon JY, Nguyen YT, et al. 10-Gingerol Targets Lipid Rafts Associated PI3K/Akt Signaling in Radio-Resistant Triple Negative Breast Cancer Cells. Molecules 2020;25:3164. [Crossref] [PubMed]
- Kiptiyah K, Widodo W, Ciptadi G, et al. 10-Gingerol as an inducer of apoptosis through HTR1A in cumulus cells: In-vitro and in-silico studies. J Taibah Univ Med Sci 2017;12:397-406. [Crossref] [PubMed]
- Rivero EM, Martinez LM, Bruque CD, et al. Prognostic significance of α- and β2-adrenoceptor gene expression in breast cancer patients. Br J Clin Pharmacol 2019;85:2143-54. [Crossref] [PubMed]
- Lavon H, Matzner P, Benbenishty A, et al. Dexmedetomidine promotes metastasis in rodent models of breast, lung, and colon cancers. Br J Anaesth 2018;120:188-96. [Crossref] [PubMed]
- Castillo LF, Rivero EM, Goffin V, et al. Alpha2-adrenoceptor agonists trigger prolactin signaling in breast cancer cells. Cell Signal 2017;34:76-85. [Crossref] [PubMed]
- Zhang D, Ma Q, Wang Z, et al. β2-adrenoceptor blockage induces G1/S phase arrest and apoptosis in pancreatic cancer cells via Ras/Akt/NFκB pathway. Mol Cancer 2011;10:146. [Crossref] [PubMed]
- Kunc M, Biernat W, Senkus-Konefka E. Estrogen receptor-negative progesterone receptor-positive breast cancer - "Nobody's land" or just an artifact? Cancer Treat Rev 2018;67:78-87. [Crossref] [PubMed]
- Ghali RM, Al-Mutawa MA, Ebrahim BH, et al. Progesterone Receptor (PGR) Gene Variants Associated with Breast Cancer and Associated Features: a Case-Control Study. Pathol Oncol Res 2020;26:141-7. [Crossref] [PubMed]
- Havas KM, Milchevskaya V, Radic K, et al. Metabolic shifts in residual breast cancer drive tumor recurrence. J Clin Invest 2017;127:2091-105. [Crossref] [PubMed]
- Gonzalez-Salinas F, Rojo R, Martinez-Amador C, et al. Transcriptomic and cellular analyses of CRISPR/Cas9-mediated edition of FASN show inhibition of aggressive characteristics in breast cancer cells. Biochem Biophys Res Commun 2020;529:321-7. [Crossref] [PubMed]
- Zielinska HA, Holly JMP, Bahl A, et al. Inhibition of FASN and ERα signalling during hyperglycaemia-induced matrix-specific EMT promotes breast cancer cell invasion via a caveolin-1-dependent mechanism. Cancer Lett 2018;419:187-202. [Crossref] [PubMed]
- Zhang Z, Yi P, Tu C, et al. Curcumin Inhibits ERK/c-Jun Expressions and Phosphorylation against Endometrial Carcinoma. Biomed Res Int 2019;2019:8912961. [Crossref] [PubMed]
- Mishra A, Bharti AC, Saluja D, et al. Transactivation and expression patterns of Jun and Fos/AP-1 super-family proteins in human oral cancer. Int J Cancer 2010;126:819-29. [PubMed]


