
Assessment of POLE and POLD1 mutations as prognosis and immunotherapy biomarkers for stomach adenocarcinoma
Introduction
DNA polymerases ε and δ, the catalytic subunits of which are encoded by the POLE and POLD1 genes respectively (1), are essential for proofreading and fidelity in DNA replication of the leading and lagging strands respectively (2-5). The fidelity of replication relies on three error avoidance mechanisms acting in series: nucleotide selectivity of replicative DNA polymerases, 3’-5’ exonucleolytic proofreading, and post-replicative DNA mismatch repair (MMR) (6,7). The proper functioning of POLE and POLD1 genes is essential to suppress gene mutations and tumorigenesis (8,9). Mutations in POLE and POLD1 genes have been associated with high tumor mutational load (TMB) (10), prognosis (11-13), and clinical benefits of immunotherapy (14-17) in many cancer types, including endometrial cancer (EC), colorectal cancer (CRC), non-small cell lung cancer (NSCLC) and glioblastoma. However, the status of POLE/POLD1 gene mutations and its clinical significance in stomach adenocarcinoma (STAD) remain unclear.
Immunotherapies based on immune checkpoint inhibitors (ICIs) have achieved remarkable clinical success and gained regulatory approval in many advanced cancers, including STAD (18-20). Gastric cancer is one of the most commonly diagnosed cancers, ranking fifth for incidence and fourth for mortality globally, and the prevalence is markedly higher in East Asia region (21). In China, it is the second most common malignancy and the third leading cause of cancer-related death, with an estimated of 403,000 new gastric cancer cases and 291,000 cancer deaths occurring in 2015 (22). STAD is the most common subtype of gastric cancer, accounting for more than 90% (23). Nowadays, PD-L1 expression, microsatellite instability (MSI)-H/deficient mismatch repair (dMMR) and TMB have been approved by the US Food and Drug Administration (FDA) as biomarkers for ICIs in the treatment of certain cancers (24-26). In addition, aneuploidy (27), Epstein-Barr virus (EBV) (28), T-cell inflamed gene profiling and interferon-γ gene signature (29,30), and circulating tumor DNA (ctDNA) (31,32) have also been reported as possible biomarkers to predict clinical outcomes of ICIs immunotherapy. Nevertheless, the assessment of these biomarkers has not reached a unified standard, and the multiplicity of these biomarkers makes us face selection difficulties in clinic. Moreover, responses to ICIs also do not fully correlate with any of these existing biomarkers (19,20), and only limited success in a minority of STAD patients has been reported in several clinical trials, with objective response rates (ORRs) of only 10–20% (20,30,33). To improve the efficacy of ICIs for STAD patients, the identification of novel accurate predictive biomarkers is becoming increasingly urgent and challenging.
In this study, we comprehensively analyzed the variations of POLE and POLD1 genes in STAD patients, examined the impacts of these mutations on the prognosis of STAD patients, and we further explored the relationship between POLE and POLD1 gene mutations and current immunotherapy biomarkers, including PD-L1 expression, TMB, aneuploidy, MSI, dMMR, and the tumor microenvironment (TME). Our research demonstrated the possibility of POLE/POLD1 mutations as novel precise biomarkers for the overall prognosis and patient selection of ICIs immunotherapy in STAD patients. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1601/rc).
Methods
Patient selection
The study population consisted of 613 STAD patients selected and retrieved from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov), Guo et al. (34) and Kakiuchi et al. (35) (Figure S1) for comprehensive integrated analysis. The cohort was obtained after removing duplicates and non-mutation information samples from cBioPortal database (https://www.cbioportal.org) (36,37). All 613 samples were summarized for POLE/POLD1 mutations and clinical characteristics, from which a total of 360 samples with both somatic mutation, mRNA expression profiling, and copy number alteration data were screened for prognostic and immunotherapy biomarker analyses. POLE/POLD1 mutations were defined as all mutations in coding regions except synonymous and intron mutations, including missense mutations, nonsense mutations, indels and splice mutations. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was deemed exempt from institutional board approval and patient informed consent was waived, due to the retrospective nature and publicly available data source of the study.
Demographic and clinicopathological variables
Baseline demographic and clinicopathological variables were treated as either categorical (e.g., grade, stage, sex) or continuous (age) as appropriate. By comparing the relationship between POLE/POLD1 mutations and these variables, the preference of these mutations was discussed.
TME analysis
We analyzed the mRNA expression levels of some cytotoxic T-cell markers (IFNG, GZMA, GZMB and GNLY) and effector cytokines (CXCL9, CXCL10 and STAT1) (12,38) through RNA-seq data (in FPKM format) to evaluate the immune activation status in STAD patients with POLE/POLD1 mutations. In addition, according to the mRNA expression level of PD-L1 and the degree of lymphocytes infiltration in the tumor, we classified the tumor immune microenvironment of POLE/POLD1-mutant STADs (39,40). The mRNA expression level of CD8A was used to indicate the presence or absence of tumor-infiltrating lymphocytes (TILs), and positive PD-L1 and CD8A were defined as above-median expression.
Immunotherapy biomarkers analysis
Currently, the biomarkers used to predict the efficacy of ICIs therapy mainly include PD-L1 expression, TMB, MSI and MMR status. Moreover, tumor aneuploidy is also considered to be negatively related to patient response to immunotherapy (27). Therefore, we analyzed the relationship between these biomarkers and POLE/POLD1 mutations to explore the possibility of using POLE/POLD1 mutations to predict the efficacy of STAD immunotherapy.
TMB was calculated with the total number of nonsynonymous somatic mutations in coding sequence (CDS). All single nucleotide variants (SNVs) and indels were included. The expression of PD-L1 and MMR-related genes were evaluated through RNA-seq data (in FPKM format). Aneuploidy scores were obtained by calculating the sum of the number of chromosome arms that were amplified or deleted. MSI status had previously been determined in the cohort by standard techniques. The effects of POLE/POLD1 mutations on these biomarkers were then comprehensively assessed.
Statistical analysis
We used the nonparametric Mann-Whitney test and Kruskal-Wallis test for all comparisons of continuous data, and analyzed the correlation between variables by Pearson rho. Categorical variables were compared using the Fisher exact test. For the analysis of the association of POLE/POLD1 mutations with clinical outcomes, survival curves were plotted using the Kaplan-Meier method and compared by the log-rank test. The OS and PFS data evaluated by Response Evaluation Criteria in Solid Tumors, version 1.1 (RECISTv1.1) were available for 341 and 343 patients, respectively. PFS was defined as the time from treatment to the date of disease progression or death from any cause. OS was defined as the time from treatment to the date of death from any cause. All statistical tests were two-sided, and p value under 0.05 was considered statistically significant. Except where indicated, statistical tests were unadjusted. Statistical analyses were performed using SPSS 25.0 and Prism 8.0 (GraphPad, USA).
Results
POLE/POLD1 mutations and STAD patient characteristics
The status of POLE/POLD1 mutations in STAD patients is summarized in Table S1. Across all 613 STAD patients, the mutation frequencies of POLE and POLD1 genes were 6.04% (37 of 613) and 2.77% (17 of 613), respectively. There were 5 patients (0.82%) had both POLE and POLD1 mutations. Overall, POLE/POLD1 mutations were detected in 49 STAD patients (7.99%), irrespective of mutation site. Among the 360 samples selected, 31 cases had POLE/POLD1 mutations, of which 24 cases had POLE mutations, 10 cases had POLD1 mutations, and 3 cases had both POLE and POLD1 mutations (Figure S2). All mutations, including P286 (41,42) and V411 (43), occurred only once except for c.347delC (mutated in 2 patients), suggesting that no hotspot mutations exist in both POLE and POLD1 genes in STAD patients. All mutations of POLE/POLD1 were distributed throughout the coding regions of the genes, including the splice regions, exonuclease domains, polymerase domains and other regions. The software, Sorts Intolerant From Tolerant (SIFT) (44) and Polymorphism Phenotyping v2 (PolyPen-2) (45) were used to predict possible impact of gene mutations on the structure and function of a protein. The lower the SIFT score, the higher the pathogenicity of the mutation, while PolyPen-2 is completely the opposite. Mutations with scores ranging from 0 to 0.05 and 0.957 to 1 are considered to be damaging, respectively. The variation was judged as a deleterious mutation when both predictions of these two software were consistent. Due to most frameshift mutations are considered damaging and may affect normal protein function, we found that most POLE/POLD1 mutations were deleterious mutations and they could occur in any regions of the entire genes, not just the exonuclease domains.
We separately analyzed those top 10 genes with high mutation frequency and statistically significant differences from wild-type samples in POLE or POLD1-mutant STAD patients (Figure S3A,B, all P-values were less than 0.001). The TTN, ARID1A, MUC16 and KMT2D genes were present in both groups. What’s more, some of these genes have been confirmed in previous studies to be related to prognosis and immunotherapy, such as LRP1B (46,47), ARID1A (48,49) and MUC16 (50,51). Most of the STADs with POLE/POLD1 mutations in the cohort also had LRP1B, ARID1A or MUC16 mutations, while most wild-type STADs did not have mutations in these three genes (Figure S4). These further prove that POLE/POLD1 mutations may be associated with prognosis and immunotherapy.
According to the status of POLE/POLD1 gene mutations, Table S2 shows the demographic and clinicopathological characteristics of this STAD cohort. Compared with wild-type STADs, patients with POLE/POLD1 mutations were correlated with older cancer onset (median age 70.5 vs. 67 years, P=0.042), and the tumors were more commonly located in the antrum anatomic subdivisions at diagnosis (48.98% vs. 31.38%, P=0.047). Besides, there were no significant differences between the POLE/POLD1-mutant group and WT group in terms of sex, stage, grade, treatment, and residual tumor.
Clinical outcomes by POLE/POLD1 mutations in STAD
We examined the association of POLE/POLD1 mutations with clinical outcomes in our entire STAD cohort. PFS of patients with POLE/POLD1-mutant STADs was not statistically significantly greater than that of other patients (Figure 1A, HR =0.47, 95% CI: 0.26–0.86, P=0.067). The same result was also observed in terms of OS (Figure 1B, HR =0.61, 95% CI: 0.35–1.05 P=0.14). Although not statistically significant, STAD patients with POLE/POLD1 mutations showed potential for favorable prognosis.
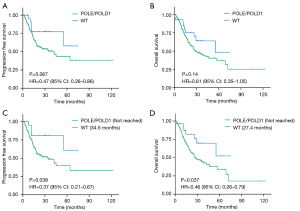
Due to the strong association of POLE/POLD1 mutations with age, especially older age onset, we speculated that their apparent prognostic effect would be most evident in related subgroup. According to research, the age of cancer onset peaks at 80 years old (22), and most of the mutant samples come from individuals under 80 (90.32%). Of 336 STAD patients under 80 years old, there were 322 and 320 patients with available PFS and OS data, respectively. In this cohort, we found a statistically significantly improved PFS (Figure 1C, not reached vs. 34.5 months, HR =0.37, 95% CI: 0.21–0.67, P=0.039) and OS (Figure 1D, not reached vs. 27.4 months, HR =0.46, 95% CI: 0.26–0.79, P=0.037) in POLE/POLD1-mutant group. As a result, POLE/POLD1 mutations can be used as prognostic markers for patients with STAD, especially in those aged 80 and younger.
POLE/POLD1 mutations increase PD-L1 expression and facilitate an adaptive immune resistance TME
Immune checkpoints PD-L1 (CD274) expression is one of the earliest biomarkers developed to enrich the population that are sensitive to PD-1/PD-L1 targeted immunotherapy in cancer patients (18,30,52), and it has been reported that it may be insufficient to be used as a stand-alone biomarker to predict the benefit of ICIs (20). To explore the potential use of POLE/POLD1 in clinical benefit of immunotherapy in STAD, we evaluated the relationship between POLE/POLD1 mutations and the mRNA expression level of PD-L1 in these patients. As illustrated in Figure 2A, POLE/POLD1-mutant STADs display significant upregulation of the expression level of PD-L1 mRNA than wild-type group (Figure 2A, P=0.0072), implying that POLE/POLD1 mutations are correlated with higher PD-L1 mRNA expression.
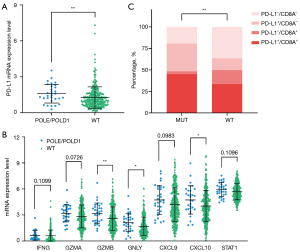
The previous study has shown that POLE mutated ECs harbor higher neoantigen loads and stimulate more potent cytotoxicity (53). To find out if POLE/POLD1 variations lead to similar response in STAD patients, we further checked the TME in POLE/POLD1-mutant STAD patients. Compared with POLE/POLD1 wild-type group, the expression of some cytotoxic T-cell markers (IFNG, GZMA, GZMB and GNLY) and effector cytokines (CXCL9, CXCL10 and STAT1) (12,38) were slightly up-regulated in STAD patients with POLE/POLD1 mutations (Figure 2B). Collectively, these data indicate the presence of a potential preexisting TILs and antitumor immunity in STAD patients with POLE/POLD1 mutations.
Based on the mRNA expression level of PD-L1 and TILs (the presence or absence of TILs), four different types of TMEs have been proposed: TME immune type (TMIT) I (high PD-L1/high CD8A), II (low PD-L1/low CD8A), III (high PD-L1/low CD8A), and IV (low PD-L1/high CD8A) (39,40). In this study, we found that the POLE/POLD1-mutant group displayed a higher proportion of dual positive PD-L1 and CD8A (PD-L1+/CD8A+) than the wild-type group (Figure 2C, 45.16% vs. 33.43%, P=0.004). This suggests that POLE/POLD1-mutant STAD patients exhibit TMIT I, a type of adaptive immune resistant TME defined as high PD-L1 expression and the presence of CD8A+ cytotoxic T lymphocytes (39), which is likely a good predictive factor for the response to anti-PD-1/PD-L1 therapy.
Higher TMB in STAD Patients with POLE/POLD1 Mutations
TMB is measured by the total number of mutations in the coding region of the tumor cell genome and has been used as an effective indicator for response prediction in immunotherapy of many cancer types (54,55). STAD patients with POLE/POLD1 mutations in the cohort exhibited a much higher number of nonsynonymous somatic mutations than wild-type patients (Figure 3A, 1,262 vs. 105, P<0.001). Consistent with the previous reports in other cancers (56,57), the TMB of MSI-H samples in STAD was significantly higher than that of MSI-L and MSS samples (Figure 3B, P<0.001). Although there was no significant difference between hypermutator phenotypes caused by POLE and POLD1 mutations (P=0.6756), the number of nonsynonymous somatic mutations in patients with POLE mutations was higher than that in MSI-H patients (MSI-H excludes samples with POLE/POLD1 mutations here; Figure 3C, 1,271 vs. 954, P=0.0168), while not the same case in POLD1-mutant group (Figure 3C, P=0.3586), suggesting MMR inactivation in the context of POLE mutations may result in a synergistic increase of mutation rate.
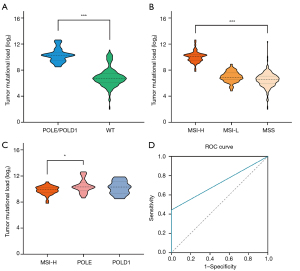
To evaluate the accuracy of using POLE/POLD1 mutations to predict the level of TMB in STAD patients, 10 mutations/Mb was used as the cut-off value for TMB as reported in most studies (58,59). The positive rate of TMB-H (>10/Mb) among POLE/POLD1-mutant patients was 100% (31/31), while that of wild-type patients was 11.85% (39/329), P<0.001 (Fisher exact test). The presence of high TMB in all types of POLE/POLD1 mutations, including indels, missense and splicing mutations, suggests that high TMB is independent of a particular mutation type, and further supports the idea that clinically significant POLE/POLD1 mutations are indeed distributed across all coding regions of the full genes.
Furthermore, to determine the sensitivity and specificity of POLE/POLD1 mutations for predicting TMB, ROC curve analysis was performed using TMB>10/Mb as the standard. The area under the ROC curve (AUC) of POLE/POLD1 mutations was 0.721, 95% CI: 0.642–0.801, with a sensitivity of 44.3% and a specificity of 100% (Figure 3D, P<0.001).
The aneuploidy score is significant lower in POLE/POLD1-mutant population
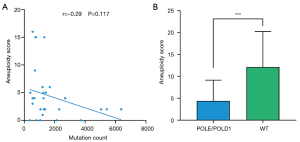
Aneuploidy, also known as somatic copy number alterations (SCNAs), is widespread in cancer and correlates with markers of immune evasion and reduced response to immunotherapy (27,60). We further explored the status of aneuploidy in POLE/POLD1-mutant STADs. As in previous research, we found that in hypermutated tumors caused by POLE/POLD1 mutations, the aneuploidy score showed a weak negative correlation with the count of mutation (Figure 4A, r=–0.29, P=0.117). Further detailed analysis found that the aneuploidy score of the POLE/POLD1-mutant group was significantly lower than that of the wild-type group (Figure 4B, P<0.001). These data validate the conclusion that TMB is much higher in STAD patients with POLE/POLD1 mutations, indicating that POLE/POLD1-mutant STADs are more likely to benefit from ICIs immunotherapy.
MSI-H is more common in STAD patients with POLE/POLD1 mutations
Multiple studies have previously shown contradictive data on the relationship between POLE/POLD1 mutations and MSI, including most POLE-mutant CRCs and ECs are hypermutant and MSS (14,61), while POLD1 mutations are correlated with higher MSI in most tumors (62). Recently, another study demonstrated that 74% POLE/POLD1-mutant patients were MSS across 47,721 patients with different cancer types (15). MSI-H tumors have been reported to have high response rate to ICIs therapy, with ORR >50% (24,63). We examined the association of POLE/POLD1 mutations with MSI in STAD patients. Among the 360 samples of STAD patients assessed in this study, 18.06% (65/360) were positive for MSI-H, and 43.08% (28/65) had POLE/POLD1 mutations. Strikingly, we found the frequency of MSI-H in both POLE and POLD1-mutant STAD patients were significantly higher than that in the wild-type group, with 91.67% (22/24) POLE-mutant and 90% (9/10) POLD1-mutant STADs were MSI-H, respectively. Overall, there were 90.32% (28/31) POLE/POLD1-mutant STAD patients correlated with higher MSI, while only 11.25% (37/329) of wild-type STADs with MSI-H (Figure 5A, P<0.001). These imply that POLE and POLD1-mutant STAD patients exhibit unique characteristics and need to be further studied extensively.
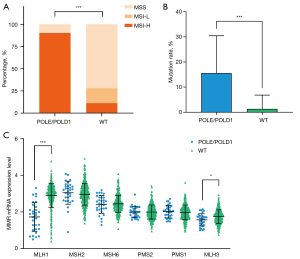
MMR deficiency is the leading cause of MSI-H (63). It has been reported that dMMR occurs in approximately 8% of early gastric cancer (stage I to stage III) and 4% of metastatic gastric cancer (63,64). The inability of the MMR proteins to function normally leads to an accumulation of errors in DNA microsatellite regions, resulting in MSI (63). We further explored the relationship between MMR-related genes (MLH1/MSH2/MSH6/PMS2/PMS1/MLH3) and POLE/POLD1, and found that the somatic mutation rate of MMR-related genes in POLE/POLD1-mutant subset was significantly higher than that of wild-type (Figure 5B, P<0.001). Combined with the above finding that most POLE/POLD1-mutant STADs are MSI-H, we hold the view that these mutations may not be passenger mutations caused by POLE/POLD1 mutations. Specifically, the POLE/POLD1-mutant group showed significantly decreased MLH1 (P<0.001) and MLH3 (P=0.0185) mRNA expression levels than that of the wild-type group (Figure 5C). Taken all these together, POLE/POLD1 mutations are correlated with higher MSI in STAD patients, and may be accompanied by higher mutation frequency of MMR-related genes and mainly decreased mRNA expression levels of MLH1 and MLH3.
Discussion
In this study, we conducted a comprehensive analysis of POLE/POLD1 gene variants in 613 STAD patients. We found the mutation frequencies of POLE and POLD1 genes in STAD are 6.04% and 2.77%, respectively. Overall, the incidence of both POLE and POLD1 gene mutations is 7.99% in STAD. And several genes believed to be associated with immunotherapy and prognosis in previous studies also had high mutation frequency in POLE/POLD1-mutant STADs. Besides, our data demonstrate that STAD patients with POLE/POLD1 mutations also exhibit favorable survival, with longer PFS and OS, especially in patients aged under 80. More importantly, the subset of STAD with POLE/POLD1 mutations displayed substantially higher PD-L1 expression, TMB and MSI, lower aneuploidy score and presented a higher proportion of Type I TME.
The frequencies of POLE and POLD1 gene mutations in STAD patients exhibited a unique pattern. The mutation frequency of POLE gene in STAD is lower than that in EC (7–12%) (14) and higher than that in CRC (1–2%) (17). Interestingly, the frequency of POLD1 mutations in STAD is higher than previously reported that POLD1 mutations are uncommon (43). Compared with a recent study from Dr. Xu’s group, the total frequency of POLE/POLD1 mutations in STAD is lower than nonmelanoma skin cancer (16.59%), EC (14.85%) and melanoma (14.7%), and a slightly higher than CRC (7.37%) (15). And STAD patients with POLE mutations were associated with an older age of onset, which is contradict with the previous reports that POLE mutations are associated with an earlier age of onset in endometrial (65) and CRC (12). More notably, STADs with POLE/POLD1 mutations also had high incidence of LRP1B, ARID1A and MUC16 mutations. LRP1B is a tumor suppressor gene that belongs to the low-density lipoprotein (LDL) receptor gene family (66), and patients with LRP1B mutation have higher TMB in lung cancer and melanoma (46,47,67). Mutations in ARID1A and MUC16 genes are related to increased immune activity of gastrointestinal tumors (48), and OS is significantly prolonged in STADs with MUC16 mutations (50). The high correlation with these clinically important genes further indicates that POLE/POLD1 gene mutations may have important significance in multiple cancer types, particularly in STAD.
EC and CRC patients with POLE mutations show favorable prognosis (11,12), and our study on the role of POLE/POLD1 mutations in STAD also obtained the same results, particularly in patients under 80 years old. Combined with what we have observed, despite the upregulation of PD-L1 mRNA and a higher proportion of PD-L1+/CD8A+ TILs in STAD subset with POLE/POLD1 mutations, the expression levels of some cytotoxic markers and effector cytokines still slightly increased, illustrating that the degree of adaptive immune resistance may be insufficient to fully suppress cytotoxicity in POLE/POLD1-mutant STADs (68), which may also be related to the better prognosis of STAD with POLE/POLD1 mutations. Besides, POLE/POLD1 positive STADs that were neither MSI-H nor PD-L1 highly expressed accounted for only a small portion (6.45%, 2 of 31), which proves that POLE/POLD1 genetic testing is helpful for treatment options of STAD patients. Moreover, STAD patients with POLE/POLD1 mutations exhibit biologically distinct characteristics. The TMB in the POLE-mutant population was even higher than that of MSI-H, suggesting STADs with POLE mutations may harbor more neoantigens and be more immunogenic. Although the analysis showed that the TMB of STAD with POLE/POLD1 mutations was not significantly different from that of MSI-H samples, this may be due to the relatively small impact of POLD1 mutations on the overall number of mutations. As dramatic ICIs responses occur only in a minority of STAD patients, and most biomarkers that have been developed and used to predict ICIs outcomes are not sufficient, therefore, the prominence of all features among POLE/POLD1-mutant STADs, including upregulation of immune checkpoint PD-L1, increased mutational load, lower aneuploidy score, and higher MSI-H percentage, implying that POLE/POLD1 mutations can be independent risk factors to predict beneficial outcome from ICIs therapy in STAD.
However, our study has some limitations. Firstly, given the retrospective nature and a relatively small population of this study, the conclusions obtained need to be verified by a large-scale prospective analysis. Secondly, the rarity and small number of POLE/POLD1-mutant STADs among the cohort warrant our findings to be interpreted with caution. Thirdly, most mutations occur in coding regions outside the polymerase and exonuclease domains, making it impossible for us to assess whether the effects of different regions within POLE/POLD1 genes are different. Fourthly, RNA-seq data rather than immunohistochemical data are used to evaluate the expression levels, and the effect of translation level on protein expression cannot be predicted. Fifthly, we did not conduct a specific analysis of the subgroup of metastatic or inoperable STADs, which is more likely to receive immunotherapy in clinical practice. Finally, the clinical outcomes of immunotherapy in these STAD patients have not been conducted in this subset, and the clinical significance of POLE/POLD1 mutations needs further study.
Considering the lack of hotspot mutations in POLE and POLD1 genes in STAD, the whole exon or even the whole gene level mutations need to be comprehensively analyzed, which makes high-throughput genetic sequencing (NGS) the ideal method to perform in the current clinical setting. All together, we propose that it is beneficial to perform genetic testing on every STAD patient before making a treatment plan, as a large amount of information can be obtained from precious patient sample, including the mutation status of the target genes (including POLE/POLD1), TMB and MSI status. In addition, germline mutations of POLE/POLD1 genes are also available through genetic testing, which can help determine whether the patient has a related genetic syndrome (69). All these data are integrated and will provide more useful and accurate information for refining risk stratification and predicting the outcomes of immunotherapy treatment in STAD.
To the best of our knowledge, this is the first study to comprehensively analyze POLE/POLD1 mutations and their correlation with current biomarkers of immunotherapy in STAD patients. Collectively, these data reveal POLE/POLD1 mutations may be very promising novel biomarkers for the risk stratification and the screening of ICIs therapy candidates in STAD. We recommend that POLE/POLD1 mutations, either alone or combined with other biomarkers, should be tested in all clinical research and practice of STAD patients. However, further studies are needed to better understand the clinical significance of POLE/POLD1 mutations as prognostic markers and predictive biomarkers for ICIs immunotherapy in STAD, which will lay a solid foundation for subsequent clinical trials and translational medicine research.
Acknowledgments
Funding: This work was supported by grants from Tianjin Cancer Hospital College-level Research Seed Fund [1810 to M Zhu], the National Natural Science Foundation of China [81700183 to L Zhang], the Second Level Candidates of 131 Innovative Talents Training Project of Tianjin [2018 to L Zhang].
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1601/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1601/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was deemed exempt from institutional board approval and patient informed consent was waived, due to the retrospective nature and publicly available data source of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Barbari SR, Shcherbakova PV. Replicative DNA polymerase defects in human cancers: Consequences, mechanisms, and implications for therapy. DNA Repair (Amst) 2017;56:16-25. [Crossref] [PubMed]
- Burgers PMJ, Kunkel TA. Eukaryotic DNA Replication Fork. Annu Rev Biochem 2017;86:417-38. [Crossref] [PubMed]
- Garbacz MA, Lujan SA, Burkholder AB, et al. Evidence that DNA polymerase δ contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat Commun 2018;9:858. [Crossref] [PubMed]
- Yeeles JTP, Janska A, Early A, et al. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol Cell 2017;65:105-16. [Crossref] [PubMed]
- Jain R, Aggarwal AK, Rechkoblit O. Eukaryotic DNA polymerases. Curr Opin Struct Biol 2018;53:77-87. [Crossref] [PubMed]
- Bębenek A, Ziuzia-Graczyk I. Fidelity of DNA replication-a matter of proofreading. Curr Genet 2018;64:985-96. [Crossref] [PubMed]
- Castellucci E, He T, Goldstein DY, et al. DNA Polymerase ɛ Deficiency Leading to an Ultramutator Phenotype: A Novel Clinically Relevant Entity. Oncologist 2017;22:497-502. [Crossref] [PubMed]
- Bellelli R, Borel V, Logan C, et al. Polε Instability Drives Replication Stress, Abnormal Development, and Tumorigenesis. Mol Cell 2018;70:707-721.e7. [Crossref] [PubMed]
- Mertz TM, Harcy V, Roberts SA. Risks at the DNA Replication Fork: Effects upon Carcinogenesis and Tumor Heterogeneity. Genes (Basel) 2017;8:46. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Church DN, Stelloo E, Nout RA, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst 2015;107:402. [Crossref] [PubMed]
- Domingo E, Freeman-Mills L, Rayner E, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol 2016;1:207-16. [Crossref] [PubMed]
- Erson-Omay EZ, Çağlayan AO, Schultz N, et al. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol 2015;17:1356-64. [Crossref] [PubMed]
- van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE Proofreading Mutations Elicit an Antitumor Immune Response in Endometrial Cancer. Clin Cancer Res 2015;21:3347-55. [Crossref] [PubMed]
- Wang F, Zhao Q, Wang YN, et al. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol 2019;5:1504-6. [Crossref] [PubMed]
- Song Z, Cheng G, Xu C, et al. Clinicopathological characteristics of POLE mutation in patients with non-small-cell lung cancer. Lung Cancer 2018;118:57-61. [Crossref] [PubMed]
- Gong J, Wang C, Lee PP, et al. Response to PD-1 Blockade in Microsatellite Stable Metastatic Colorectal Cancer Harboring a POLE Mutation. J Natl Compr Canc Netw 2017;15:142-7. [Crossref] [PubMed]
- Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:2461-71. [Crossref] [PubMed]
- Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric Cancer 2020;23:565-78. [Crossref] [PubMed]
- Sundar R, Smyth EC, Peng S, et al. Predictive Biomarkers of Immune Checkpoint Inhibition in Gastroesophageal Cancers. Front Oncol 2020;10:763. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Zheng RS, Sun KX, Zhang SW, et al. Report of cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi 2019;41:19-28. [PubMed]
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am 2013;42:211-7. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Liu J, Zhou M, Ouyang Y, et al. Identification of potential biomarkers and their clinical significance in gastric cancer using bioinformatics analysis methods. PeerJ 2020;8:e9174. [Crossref] [PubMed]
- Chung HC, Ros W, Delord JP, et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Cervical Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol 2019;37:1470-8. [Crossref] [PubMed]
- Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355:eaaf8399. [Crossref] [PubMed]
- Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449-58. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Fuchs CS, Doi T, Jang RW, et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol 2018;4:e180013. [Crossref] [PubMed]
- Cabel L, Proudhon C, Romano E, et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol 2018;15:639-50. [Crossref] [PubMed]
- Jin Y, Chen DL, Wang F, et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer 2020;19:154. [Crossref] [PubMed]
- Chen LT, Satoh T, Ryu MH, et al. A phase 3 study of nivolumab in previously treated advanced gastric or gastroesophageal junction cancer (ATTRACTION-2): 2-year update data. Gastric Cancer 2020;23:510-9. [Crossref] [PubMed]
- Guo YA, Chang MM, Huang W, et al. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat Commun 2018;9:1520. [Crossref] [PubMed]
- Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583-7. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Ock CY, Keam B, Kim S, et al. Pan-Cancer Immunogenomic Perspective on the Tumor Microenvironment Based on PD-L1 and CD8 T-Cell Infiltration. Clin Cancer Res 2016;22:2261-70. [Crossref] [PubMed]
- Teng MW, Ngiow SF, Ribas A, et al. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res 2015;75:2139-45. [Crossref] [PubMed]
- Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res 2014;74:1895-901. [Crossref] [PubMed]
- Parkash V, Kulkarni Y, Ter Beek J, et al. Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase ε. Nat Commun 2019;10:373. [Crossref] [PubMed]
- Church DN, Briggs SE, Palles C, et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet 2013;22:2820-8. [Crossref] [PubMed]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073-81. [Crossref] [PubMed]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248-9. [Crossref] [PubMed]
- Johnson DB, Frampton GM, Rioth MJ, et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 2016;4:959-67. [Crossref] [PubMed]
- Lan S, Li H, Liu Y, et al. Somatic mutation of LRP1B is associated with tumor mutational burden in patients with lung cancer. Lung Cancer 2019;132:154-6. [Crossref] [PubMed]
- Li L, Li M, Jiang Z, et al. ARID1A Mutations Are Associated with Increased Immune Activity in Gastrointestinal Cancer. Cells 2019;8:678. [Crossref] [PubMed]
- Shen J, Ju Z, Zhao W, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med 2018;24:556-62. [Crossref] [PubMed]
- Li X, Pasche B, Zhang W, et al. Association of MUC16 Mutation With Tumor Mutation Load and Outcomes in Patients With Gastric Cancer. JAMA Oncol 2018;4:1691-8. [Crossref] [PubMed]
- Zhang L, Han X, Shi Y. Association of MUC16 Mutation With Response to Immune Checkpoint Inhibitors in Solid Tumors. JAMA Netw Open 2020;3:e2013201. [Crossref] [PubMed]
- Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014;20:5064-74. [Crossref] [PubMed]
- Howitt BE, Shukla SA, Sholl LM, et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol 2015;1:1319-23. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Castle JC, Uduman M, Pabla S, et al. Mutation-Derived Neoantigens for Cancer Immunotherapy. Front Immunol 2019;10:1856. [Crossref] [PubMed]
- Zang YS, Dai C, Xu X, et al. Comprehensive analysis of potential immunotherapy genomic biomarkers in 1000 Chinese patients with cancer. Cancer Med 2019;8:4699-708. [Crossref] [PubMed]
- Borghaei H, Hellmann MD, Paz-Ares LG, et al. Nivolumab (Nivo)+ platinum-doublet chemotherapy (Chemo) vs. chemo as first-line (1L) treatment (Tx) for advanced non-small cell lung cancer (NSCLC) with< 1% tumor PD-L1 expression: Results from CheckMate 227. ASCO Annual Meeting; 2018.
- Mishima S, Kawazoe A, Nakamura Y, et al. Clinicopathological and molecular features of responders to nivolumab for patients with advanced gastric cancer. J Immunother Cancer 2019;7:24. [Crossref] [PubMed]
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676-689.e3. [Crossref] [PubMed]
- Guerra J, Pinto C, Pinto D, et al. POLE somatic mutations in advanced colorectal cancer. Cancer Med 2017;6:2966-71. [Crossref] [PubMed]
- Fabrizio DA, George TJ Jr, Dunne RF, et al. Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J Gastrointest Oncol 2018;9:610-7. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Pietrantonio F, Miceli R, Raimondi A, et al. Individual Patient Data Meta-Analysis of the Value of Microsatellite Instability As a Biomarker in Gastric Cancer. J Clin Oncol 2019;37:3392-400. [Crossref] [PubMed]
- Billingsley CC, Cohn DE, Mutch DG, et al. Polymerase ɛ (POLE) mutations in endometrial cancer: clinical outcomes and implications for Lynch syndrome testing. Cancer 2015;121:386-94. [Crossref] [PubMed]
- Liu CX, Li Y, Obermoeller-McCormick LM, et al. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem 2001;276:28889-96. [Crossref] [PubMed]
- Chen H, Chong W, Wu Q, et al. Association of LRP1B Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front Immunol 2019;10:1113. [Crossref] [PubMed]
- Speiser DE, Utzschneider DT, Oberle SG, et al. T cell differentiation in chronic infection and cancer: functional adaptation or exhaustion? Nat Rev Immunol 2014;14:768-74. [Crossref] [PubMed]
- Buchanan DD, Stewart JR, Clendenning M, et al. Risk of colorectal cancer for carriers of a germ-line mutation in POLE or POLD1. Genet Med 2018;20:890-5. Erratum in: Genet Med 2018 Feb 01. [Crossref] [PubMed]


