
Cleavage factor Im 25 as a potential biomarker for prognosis of colorectal cancer
Introduction
According to global cancer statistics in 2018, there were about 1.8 million new cases of colorectal cancer (CRC) and 861,663 new deaths (1). Approximately one third of CRC patients have distant metastasis, most commonly in the liver, and the 5-year overall survival rate is only 12.5% (2,3). Only 15% of cases in CRC patients with distant metastasis are considered resectable, and over 50% relapse within 2 years (4,5). Because the molecular mechanisms of CRC are still not well understood, existing treatment methods are insufficient for metastatic patients (6). Therefore, it is necessary to investigate the molecular pathogenesis underlying the development and progression of CRC to identify novel therapeutic targets.
Cleavage factor Im 25 (CFIm25) is a key regulator of Alternative polyadenylation (APA), and is a 25 kDa subunit of cleavage factor Im, encoded by gene NUDT21; its core component is the typical Nudix structure, which can bind the UGUA element of pre-mRNA and promote the formation of an APA initiation complex by conferring the surface residue with a negative charge, and then playing the role of a cleavage factor (7,8). Alternative polyadenylation is a phenomenon in which the pre-mRNA is cleaved at different polyadenylation sites in the 3' end untranslated region (3'UTR) and added to an untemplated poly(A) tract, causing the expression of substantial isoforms of mRNA with differing 3'UTR lengths, which are controlled by the downstream element (GUGU) (9), There are many sequence elements regulated by miRNA in the 3'UTR (10); therefore, APA can promote escape from miRNA inhibition by shortening the 3'UTR of genes and increasing gene expression, thus affecting the progress and prognosis of seven types of carcinoma (11). The CFIm25 protein has multiple polyadenylation sites in its 3'UTR and can regulate APA site selection. The decreased expression of CFIm25 protein causes the poly(A) site to transfer from a distal location to proximal a proximal one, resulting in a shortened mRNA 3'UTR (12). Weng et al. found that the decreased expression of CFIm25 can result in the loss of miRNA sites by shortening the 3'UTR of target genes, causing these genes to escape inhibition by miRNA and become overexpressed, thereby promoting the proliferation of fibroblasts; conversely, overexpression of CFIm25 can lengthen the 3'UTR of target genes, reducing their expression and inhibiting the proliferation of fibroblasts (13). CFIm25 is also reported to participate in the progression and prognosis of carcinoma. CFIm25 can inhibit the growth of non-small cell lung cancer cells (14). Furthermore, CFIm25 not only plays a role in solid tumors but also in leukemia (15). Whether CFIm25 is associated with survival in patients with CRC remains unclear. Therefore, we examined the expression of CFIm25 in CRC to explore its role. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1441).
Methods
Patient specimens
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Jiangmen Central Hospital (2021-8), and individual consent for this retrospective analysis was waived.
A total of 363 cases of CRC (209 men and 154 women) were collected during surgical resection at Jiangmen Central Hospital between 2010 and 2011. The mean age was 60. The follow-up of all patients was censored on February 2021. The eligibility criteria of the present study were as follows: CRC was the only primary tumor; no secondary primary tumor; no history of chemotherapy, radiotherapy, or surgery before the resection of CRC; the tissue was suitable for immunohistochemical examination. All cases were classified by an attending pathologist according to the World Health Organization (WHO) Classification of Tumors of the Digestive System. Tumor, node, and metastasis (TNM) staging was performed according to the American Joint Committee on Cancer (AJCC) cancer staging manual (Eighth Edition).
Tissue microarray (TMA) and immunohistochemistry (IHC)
All CRC tissues were fixed with 10% neutral buffered formalin (NBF) at room temperature for 48 h. TMAs were generated from paraffin-embedded tissue blocks according to the standard EnVision Tissue Microarray protocol. Three samples with a diameter of 1.5 mm were punched from the carcinoma areas of each sample and transferred into the TMA. Three-micrometer TMA sections were used for the IHC sections. After dewaxing with xylene and rehydration in a descending alcohol series and distilled water, the sections were incubated in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity, which was carried out at room temperature. To recover the antigen, the sections were immersed in citric acid buffer solution (pH 9.0) and pressure-cooked at 100 °C for 3 min. The sections were incubated with a CFIm25 primary antibody (Mouse monoclonal antibody; OTI13H1, LSBio) at a dilution of 1:200 for 50 min at 37 °C in an incubator. They were then washed with buffer and the secondary antibody was added (undiluted; rabbit. no. K5007; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) for 30 min at 37 °C in an incubator. Staining was performed using 3,3-diaminoaniline. The sections were then counterstained with hematoxylin, dehydrated, then immersed in xylene. Finally, the sections were sealed with neutral balsam. Thyroid tissue was used as the positive control, which was sourced from paraffin-embedded tissue specimens that had been archived following pathological diagnosis. The negative control used 0.02 mol/L PBS instead of the primary antibody to incubate sections.
IHC evaluation
The Aperio AT2 (Leica Microsystems B.V., Wetzlar, Germany) scanner was used to digitally capture images of the stained sections. Two attending pathologists independently evaluated the expression of CFIm25 after imaging the stained sections. The dye strength was evaluated as negative “−”, weak “1+”, moderate “2+”, and strong “3+”; the number of positive tumor cells was defined as the percentage of all cells (0–100%). The scoring of each sample was evaluated by multiplying the percentage of positive tumor cells by dye strength, resulting in scores ranging 0–300. The average of the three points was used to calculate the TMA IHC results.
Statistical analysis
SPSS V16.0 (SPSS, Inc.) was used for statistical analysis and survival curve drawing. The associations between CFIm25 expression and clinicopathological variables were calculated using the χ2 test. The Kaplan-Meier method with log-rank test was used to analyze survival. Multivariate analyses were performed using a Cox proportional hazards model. Statistical significance was set at P<0.05. All P values were analyzed bilaterally, and the mean standard deviation was used to present all data.
Results
CFIm25 expression cut-off values
A receiver operating characteristic (ROC) curve was used to determine the optimal cut-off value for CFIm25 expression. The ROC analysis of survival status, pT stage, pN stage, pM stage, vascular invasion, and serosa invasion (Figure 1), it was found that the highest point of survival status in the ROC curve was the closest point (0.0, 1.0), with the highest sensitivity and specificity, and the best classification efficiency (area under curve, 0.607; P=0.001, Figure 1A). Taking the survival state as a state variable, 165 was defined as the cut-off value of CFIm25 expression. Values ≥165 indicate overexpression, and those <165 indicate low expression.

CFIm25 expression level and correlation with clinicopathological features in CRC patients
CFIm25 was expressed in most CRC patients. All of them are expressed in the nuclei of the tumor cells. According to the cut-off value of 165, 144 (39.7%) cases had overexpression and 219 (60.3%) cases had low expression (Figure 2). The χ2 test was used for further analysis, finding that CFIm25 expression was significantly associated with vascular invasion (P=0.000), serosal invasion (P=0.007), pT stage (P=0.016), and clinical stage (P=0.007), but not with sex, age, tumor size, nerve invasion, tumor differentiation, infiltration type, pN stage, pM stage, and relapse. All data are presented in Table 1. These results indicate that the expression of CFIm25 may be associated with the progression of CRC.
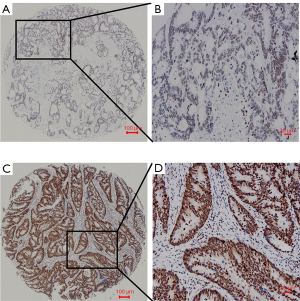
Table 1
| Variables | CFIM25 expression | |||
|---|---|---|---|---|
| All | Low, n (%) | High, n (%) | P value | |
| Gender | 0.287 | |||
| Female | 154 | 88 (57.1) | 66 (42.9) | |
| Male | 209 | 131 (62.7) | 78 (37.3) | |
| Age1 | 0.345 | |||
| ≤60 years | 183 | 106 (57.9) | 77 (42.1) | |
| >60 years | 180 | 113 (62.8) | 67 (37.2) | |
| Tumor size2 | 0.267 | |||
| ≤4 cm | 192 | 121 (63.0) | 71 (37.0) | |
| >4 cm | 171 | 98 (57.3) | 73 (42.7) | |
| Tumor differentiation | 0.149 | |||
| Well | 32 | 19 (59.4) | 13 (40.6) | |
| Moderate | 307 | 181 (59.0) | 126 (41.0) | |
| Poor | 24 | 19 (79.2) | 5 (20.8) | |
| Infiltration type | 0.494 | |||
| Infiltration | 19 | 9 (47.4) | 10 (52.6) | |
| Ulcerative | 206 | 126 (61.2) | 80 (38.8) | |
| Uplift | 138 | 84 (60.9) | 54 (39.1) | |
| pT stage | 0.016a | |||
| T1 | 10 | 4 (40.0) | 6 (60.0) | |
| T2 | 81 | 41 (50.6) | 40 (49.4) | |
| T3 | 106 | 60 (56.6) | 46 (43.4) | |
| T4 | 166 | 114 (68.7) | 52 (31.3) | |
| pN stage | 0.415 | |||
| N0 | 206 | 130 (63.1) | 76 (36.9) | |
| N1 | 103 | 57 (55.3) | 46 (44.7) | |
| N2 | 54 | 32 (59.3) | 22 (40.7) | |
| pM stage | 0.098 | |||
| M0 | 295 | 184 (62.4) | 111 (37.6) | |
| M1 | 68 | 35 (51.5) | 33 (48.5) | |
| Clinical stage | 0.007a | |||
| I | 66 | 34 (51.5) | 32 (48.5) | |
| II | 123 | 89 (72.4) | 34 (27.6) | |
| III | 106 | 61 (57.5) | 45 (42.5) | |
| IV | 68 | 35 (51.5) | 33 (48.5) | |
| Relapse | 0.059 | |||
| No | 287 | 166 (57.8) | 121 (42.2) | |
| Yes | 76 | 53 (69.7) | 23 (30.3) | |
| Vascular invasion | 0.000b | |||
| No | 255 | 139 (54.5) | 116 (45.5) | |
| Yes | 108 | 80 (74.1) | 28 (25.9) | |
| Nerve invasion | 0.488 | |||
| No | 224 | 132 (58.9) | 92 (41.1) | |
| Yes | 139 | 87 (62.6) | 52 (37.4) | |
| Serosa invasion | 0.007a | |||
| No | 195 | 105 (53.8) | 90 (46.2) | |
| Yes | 168 | 114 (67.9) | 54 (32.1) | |
Age1: median; Tumor size2: median; aP<0.05; bP<0.001. CFIm25, cleavage factor Im 25; CRC, colorectal cancer.
Association between clinicopathological features and survival status of patients with CRC
The univariate survival analysis of this data set revealed that the median survival time of CRC onset was significantly different before and after 60 years of age (P=0.012). The OS values of patients with stage T1, T2, T3, and T4 were 119.50, 115.20, 106.28, and 80.44 months, respectively (P=0.000, Figure 3A). Figure 3B shows that as the N stage progressed, the OS values of patients with CRC differed significantly (P=0.000; Figure 3B). Compared with the 105.33 months of stage M0, the OS of stage M1 was 58.79 months, about half of that of stage M0 (P=0.000; Figure 3C). The OS values of CRC patients with stage I, II, III, IV were 118.45, 108.24, 92.25, and 58.79 months respectively (P=0.000, Figure 3D). Figure 3E shows that the OS was shorter in patients who had suffered relapse than in those who had not (P=0.000; Figure 3E). The OS of patients with well-differentiated, moderately differentiated, and poorly differentiated tumors were 113.24, 97.67, and 67.88 months, respectively (P=0.005; Figure 3F). Figure 3G shows that the OS of patients with vascular invasion was shorter than that of patients without vascular invasion (P=0.000; Figure 3G). In patients with neural invasion, the OS was 32.5 months shorter than in patients without neural invasion (P=0.000; Figure 3H). While in patients with serosal invasion, the survival time was only 80.71 months and was 111.46 months in patients without it (P=0.000, Figure 3I). All data are presented in Table 2.
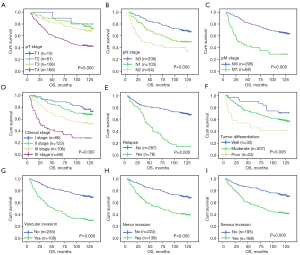
Table 2
| Variables | All cases | Mean survival (months) | Chi-square value | P valued |
|---|---|---|---|---|
| Gender | 0.915 | 0.339 | ||
| Female | 154 | 94.34±3.93 | ||
| Male | 209 | 99.33±3.22 | ||
| Age1, years | 6.26 | 0.012a | ||
| ≤60 | 183 | 102.32±3.41 | ||
| >60 | 180 | 91.87±3.62 | ||
| Tumor size2 | 0.122 | 0.727 | ||
| ≤4 cm | 192 | 99.43±3.27 | ||
| >4 cm | 171 | 94.73±3.82 | ||
| Tumor differentiation | 7.95 | 0.005b | ||
| Well | 32 | 113.24±6.06 | ||
| Moderate | 307 | 97.67±2.68 | ||
| Poor | 24 | 67.88±11.21 | ||
| Infiltration type | 1.778 | 0.182 | ||
| Infiltration | 19 | 82.67±10.56 | ||
| Ulcerative | 206 | 96.20±3.32 | ||
| Uplift | 138 | 99.42±3.95 | ||
| pT stage | 31.19 | 0.000c | ||
| T1 | 10 | 119.50±8.33 | ||
| T2 | 81 | 115.20±4.04 | ||
| T3 | 106 | 106.28±4.04 | ||
| T4 | 166 | 80.44±3.93 | ||
| pN stage | 34.66 | 0.000c | ||
| N0 | 206 | 109.05±2.77 | ||
| N1 | 103 | 88.98±4.85 | ||
| N2 | 54 | 64.87±7.15 | ||
| pM stage | 59.65 | 0.000c | ||
| M0 | 295 | 105.33±2.44 | ||
| M1 | 68 | 58.79±6.38 | ||
| Clinical stage | 52.21 | 0.000c | ||
| I | 66 | 118.45±3.81 | ||
| II | 123 | 108.24±3.37 | ||
| III | 106 | 92.25±4.65 | ||
| IV | 68 | 58.79±6.38 | ||
| Relapse | 84.67 | 0.000c | ||
| No | 287 | 106.76±2.62 | ||
| Yes | 76 | 59.44±4.52 | ||
| Vascular invasion | 64.59 | 0.000c | ||
| No | 255 | 109.61±2.54 | ||
| Yes | 108 | 68.01±4.81 | ||
| Nerve invasion | 37.26 | 0.000c | ||
| No | 224 | 109.26±2.71 | ||
| Yes | 139 | 76.76±4.28 | ||
| Serosa invasion | 36.43 | 0.000c | ||
| No | 195 | 111.46±2.85 | ||
| Yes | 168 | 80.71±3.89 | ||
| CFIm25 expression | 14.97 | 0.000c | ||
| Low | 219 | 88.53±3.38 | ||
| High | 144 | 110.69±3.32 |
Age1: median; Tumor size2: median; aP<0.05, bP<0.01, cP<0.001. dLog-rank test. CRC, colorectal cancer; CFIm25, Cleavage Factor Im 25.
Low CFIm25 expression results in a poorer prognosis
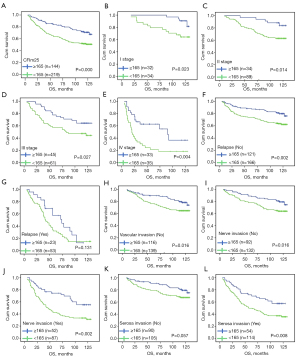
Univariate analysis showed that the OS of patients with high CFIm25 expression was significantly different from that of patients with low CFIm25 expression. The survival time of CRC patients with low CFIm25 expression was only 88.53 months, while the survival time of CRC patients with high CFIm25 expression was 110.69 months (P=0.000, Table 2, and Figure 4A). Further stratified analysis of each clinical feature showed that the expression level of CFIm25 and survival time were significantly different in different subgroups, suggesting that CFIm25 protein has clinical classification function in patients with CRC. In patients with stage I CRC, those with high CFIm25 expression had longer survival than those with low CFIm25 expression (P=0.023, Figure 4B). Likewise, similar results were found in patients with stage II, III, and IV CRC (P=0.014, Figure 4C; P=0.027, Figure 4D; P=0.004, Figure 4E). Patients with high CFIm25 protein expression in the non-recurrent CRC subgroup had a longer survival time (P=0.002, Figure 4F), whereas the efficiency of the classifier was not obvious in the relapsed subgroup (P=0.131, Figure 4G). CFim25 was also found to be a significant classifier for survival analysis in subgroups of CRC patients with no vascular invasion, no nerve invasion, and nerve invasion (P=0.016, Figure 4H; P=0.016, Figure 4I; P=0.002, Figure 4J). Stratified analysis of CFIm25 expression in the subgroup of patients with CRC and serosal invasion also revealed that high CFIm25 expression in patients with CRC and serosal invasion had a longer survival period, while the difference was not significant in the subgroup with no serosal invasion (P=0.057, Figure 4K; P=0.008, Figure 4L).
Independent prognostic factors for CRC
The Cox risk regression model was used for further analysis, and the results showed that CFIm25 is an independent prognostic factor for CRC (HR, 0.543; 95% CI: 0.372–0.792; P=0.002). In addition, age (HR, 1.455; 95% CI, 1.045–2.026; P=0.027), vascular invasion (HR, 2.240; 95% CI: 1.599–3.138; P=0.000), nerve invasion (HR, 1.827; 95% CI: 1.309–2.550; P=0.000), serosa invasion (HR, 1.470; 95% CI: 1.032–2.094; P=0.033), relapse (HR, 2.776; 95% CI: 1.965–3.922; P=0.000), clinical stage (HR, 1.597; 95% CI: 1.297–1.965; P=0.000) are the independent prognostic factor for patients with CRC. All data are presented in Table 3.
Table 3
| Variable | OS | |
|---|---|---|
| HR (95% CI) | P value | |
| Age (>60 vs. ≤60 years) | 1.455 (1.045–2.026) | 0.027a |
| Clinical stage (IV vs. III vs. II vs. I) | 1.597 (1.297–1.965) | 0.000c |
| Relapse (yes vs. no) | 2.776 (1.965–3.922) | 0.000c |
| Vascular invasion (yes vs. no) | 2.240 (1.599–3.138) | 0.000c |
| Nerve invasion (yes vs. no) | 1.827 (1.309–2.550) | 0.000c |
| Serosa invasion (yes vs. no) | 1.470 (1.032–2.094) | 0.033a |
| CFIM25 expression (high vs. low) | 0.543 (0.372–0.792) | 0.002b |
aP<0.05, bP<0.01, cP<0.001. HR, hazard ratio; OS, overall survival; CI, confidence interval; CRC, colorectal cancer.
Discussion
In recent years, the mechanism of CFIm25 protein in cancer has been widely studied (16). Previous studies have shown that the CFIm25 protein has multiple poly(A) site within 3'UTR, when CFIm25 protein decreased expression, the poly(A) site can be transferred from a distal position to a proximal one, shortening the 3'UTR of a subset of genes (12). These genes subsequently participate in various signaling pathways, cell proliferation and transformation, and the competing-endogenous RNA (ceRNA) regulatory mechanism (17). This epigenetic mechanism provides a novel direction for cancer treatment.
In this study, the expression of CFIm25 protein was detected by IHC in the tissues of patients with CRC, and the clinical data of the patients were retrospectively analyzed. We found that age, serosa invasion, vascular invasion, nerve invasion, relapse, clinical stage, and CFIm25 were independent prognostic factors for CRC. Studies have reported that serosal invasion is a predictor of gastric cancer recurrence and peritoneal metastasis (18,19). However, sufficient attention has not been paid to serosal invasion in CRC, and this study once again confirmed that serosal invasion was involved in the progression and prognosis of CRC. Therefore, this may result in a higher risk factor for CRC patients.
A previous study showed that decreased expression of CFIm25 protein is involved in cellular pluripotency reprogramming and malignant transformation through cancer-related pathways (20). In glioblastoma, the decreased expression of CFIm25 protein shortened the 3'UTR of PAK1 and overexpressed PAK1, then promote the progression of carcinoma by activating the PAK1-RAS signaling pathway (21). In lung cancer, the decreased CFIm25 expression shortens the 3'UTR of cyclinD1 and activates the cyclinD1 pathway to promote tumor cell proliferation (22). Interestingly, studies have shown that the 3'UTR of glutaminase (GLS) is also regulated by CFIm25, and the decreased CFIm25 expression can regulate the GAC:KGA isotype ratio to change GLS metabolism and provide nutrients for proliferating cells to promote the progression of carcinoma (23,24). These studies showed that decreased expression of CFIm25 protein can promote the proliferation of tumor cells. Similar results were found in our study: CFIm25 acted as a tumor suppressor gene. CRC patients with decreased or increased CFIm25 expression had shorter or longer survival times, respectively. The CFIm25 protein is an independent prognostic factor for CRC, but its molecular mechanism remains unclear. Masamha et al. performed RNA sequencing analysis of APA in gliomas, and revealed that knockdown of CFIm25 resulted in at least 1,450 genes with shorter 3'UTRs and a significant increase in the expression of some known oncogenes (25). Another study showed decreased CFIm25 expression can use the proximal poly(A) site to shorten the 3'UTR and result in the loss of miRNA binding sites, causing target genes to escape inhibition by miRNA and overexpression, promoting the proliferation of hepatoma cells (26). Tan et al. found a similar mechanism in hepatocellular carcinoma: the overexpression of CFIm25 protein can transfer the poly(A) site of PSMB2 and CXXC5 to the distal end, produce a longer subtype of 3'UTR, reduce the expression of PSMB2 and CXXC5, and finally inhibit the proliferation, metastasis, and tumorigenesis of hepatocellular carcinoma (27). In bladder cancer, the overexpression of CFIm25 elongated the 3'UTRs of ANXA2 (annexin A2) and LIMK2 (lim domain kinase 2), thus reducing the expression of these genes, leading to the inhibition of Wnt/β-catenin and NF-κB signaling pathways, and inhibiting the progression of bladder cancer (28).
Another study reported that in cervical cancer, decreased CFIm25 expression can shorten the 3'UTR of WNT10B and HMGB1, and the expression of these genes was negatively correlated with CFIm25 expression, when then activates the Wnt/β-catenin and NF-κB signaling pathways to promote carcinoma progression (29). In breast cancer, decreased expression of CFIm25 can shorten the 3'UTR of a subset of genes while causing the loss ofmiRNA target sites and promoting tumor cell proliferation (30). These findings suggest that CFIm25 plays a dual role as a tumor suppressor and cancer promoter in the development and progression of tumors. Morris et al. found that APA can greatly shorten the 3'UTR in the malignant transformation process of CRC (31). Therefore, we hypothesized that there would be a similar mechanism of action for CFIm25 in the development of CRC. The decreased expression of CFIm25 may shorten the 3'UTR of a subset of genes, causing the 3'UTR to lose its miRNA site. Then, the genes can escape the inhibition by miRNA and become overexpressed, and then activate the ceRNA pathway, which promotes the progression of CRC. The molecular mechanism of CFIm25 in CRC remains to be further explored.
In summary, this study showed that decreased expression of CFIm25 indicates a worse prognosis for CRC patients, which suggests that CFIm25 may be a novel potential biomarker for CRC in the future.
Acknowledgments
Funding: The present study was supported by the Youth Foundation of National Natural Science Foundation of China (81902420), Guangdong Esophageal Cancer Institute Science and Technology Program Project (Q201903), and the Medical Technology Foundation of Jiangmen (2017A4003).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1441
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-1441
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1441). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Jiangmen Central Hospital (2021-8) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252-71. [Crossref] [PubMed]
- Manfredi S, Lepage C, Hatem C, et al. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg 2006;244:254-9. [Crossref] [PubMed]
- de Jong MC, Pulitano C, Ribero D, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg 2009;250:440-8. [Crossref] [PubMed]
- Poston GJ, Figueras J, Giuliante F, et al. Urgent need for a new staging system in advanced colorectal cancer. J Clin Oncol 2008;26:4828-33. [Crossref] [PubMed]
- Pitroda SP, Khodarev NN, Huang L, et al. Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat Commun 2018;9:1793. [Crossref] [PubMed]
- Coseno M, Martin G, Berger C, et al. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res 2008;36:3474-83. [Crossref] [PubMed]
- Yang Q, Gilmartin GM, Doublié S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3' processing. Proc Natl Acad Sci U S A 2010;107:10062-7. [Crossref] [PubMed]
- Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 2017;18:18-30. [Crossref] [PubMed]
- Mayr C. Evolution and Biological Roles of Alternative 3'UTRs. Trends Cell Biol 2016;26:227-37. [Crossref] [PubMed]
- Xia Z, Donehower LA, Cooper TA, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3'-UTR landscape across seven tumour types. Nat Commun 2014;5:5274. [Crossref] [PubMed]
- Kubo T, Wada T, Yamaguchi Y, et al. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3'-UTRs. Nucleic Acids Res 2006;34:6264-71. [Crossref] [PubMed]
- Weng T, Ko J, Masamha CP, et al. Cleavage factor 25 deregulation contributes to pulmonary fibrosis through alternative polyadenylation. J Clin Invest 2019;129:1984-99. [Crossref] [PubMed]
- Cheng ZJ, Miao DL, Su QY, et al. THZ1 suppresses human non-small-cell lung cancer cells in vitro through interference with cancer metabolism. Acta Pharmacol Sin 2019;40:814-22. [Crossref] [PubMed]
- Zhang L, Zhang W. Knockdown of NUDT21 inhibits proliferation and promotes apoptosis of human K562 leukemia cells through ERK pathway. Cancer Manag Res 2018;10:4311-23. [Crossref] [PubMed]
- Sun X, Li J, Sun X, et al. CFIm25 in Solid Tumors: Current Research Progress. Technol Cancer Res Treat 2020;19:1533033820933969. [Crossref] [PubMed]
- Jafari Najaf Abadi MH, Shafabakhsh R, Asemi Z, et al. CFIm25 and alternative polyadenylation: Conflicting roles in cancer. Cancer Lett 2019;459:112-21. [Crossref] [PubMed]
- Roukos DH, Lorenz M, Karakostas K, et al. Pathological serosa and node-based classification accurately predicts gastric cancer recurrence risk and outcome, and determines potential and limitation of a Japanese-style extensive surgery for Western patients: a prospective with quality control 10-year follow-up study. Br J Cancer 2001;84:1602-9. [Crossref] [PubMed]
- Togano S, Yashiro M, Miki Y, et al. Microscopic distance from tumor invasion front to serosa might be a useful predictive factor for peritoneal recurrence after curative resection of T3-gastric cancer. PLoS One 2020;15:e0225958. [Crossref] [PubMed]
- Brumbaugh J, Di Stefano B, Wang X, et al. Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling. Cell 2018;172:106-120.e21. [Crossref] [PubMed]
- Chu Y, Elrod N, Wang C, et al. Nudt21 regulates the alternative polyadenylation of Pak1 and is predictive in the prognosis of glioblastoma patients. Oncogene 2019;38:4154-68. [Crossref] [PubMed]
- Huang J, Weng T, Ko J, et al. Suppression of cleavage factor Im 25 promotes the proliferation of lung cancer cells through alternative polyadenylation. Biochem Biophys Res Commun 2018;503:856-62. [Crossref] [PubMed]
- Gao CC, Xu QQ, Xiao FJ, et al. NUDT21 suppresses the growth of small cell lung cancer by modulating GLS1 splicing. Biochem Biophys Res Commun 2020;526:431-8. [Crossref] [PubMed]
- Masamha CP, Xia Z, Peart N, et al. CFIm25 regulates glutaminase alternative terminal exon definition to modulate miR-23 function. RNA 2016;22:830-8. [Crossref] [PubMed]
- Masamha CP, Xia Z, Yang J, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 2014;510:412-6. [Crossref] [PubMed]
- Sun M, Ding J, Li D, et al. NUDT21 regulates 3'-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett 2017;410:158-68. [Crossref] [PubMed]
- Tan S, Li H, Zhang W, et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene 2018;37:4887-900. [Crossref] [PubMed]
- Xiong M, Chen L, Zhou L, et al. NUDT21 inhibits bladder cancer progression through ANXA2 and LIMK2 by alternative polyadenylation. Theranostics 2019;9:7156-67. [Crossref] [PubMed]
- Xing Y, Chen L, Gu H, et al. Downregulation of NUDT21 contributes to cervical cancer progression through alternative polyadenylation. Oncogene 2021;40:2051-64. [Crossref] [PubMed]
- Kim S, Bai Y, Fan Z, et al. The microRNA target site landscape is a novel molecular feature associating alternative polyadenylation with immune evasion activity in breast cancer. Brief Bioinform 2021;22:bbaa191.
- Morris AR, Bos A, Diosdado B, et al. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res 2012;18:5256-66. [Crossref] [PubMed]


