
Clinicopathological implications of lncRNAs, immunotherapy and DNA methylation in lung squamous cell carcinoma: a narrative review
Introduction
Lung cancer is still the leading cause of cancer mortality worldwide. The most prevalent lung cancer is non-small cell lung cancer (NSCLC), accounting for 80–85 percent of cases (1,2). The histological classifications used to classify NSCLC include lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LAUD), neuroendocrine cancer, and large cell carcinoma (3). The most prevalent form of NSCLC is LUSC, with a high risk of metastasis and recurrence (4). Consequently, new therapeutic biomarkers and strategies for directing future therapies to mitigate the impact on LUSC must be studied and discovered.
The significance of non-coding RNAs (ncRNA) as essential regulators in many cellular activities have been more evident over the last decade (5). Non-protein coding RNA transcripts of >200 nucleotides long are referred to as long non-coding RNAs (6,7). While lncRNAs are typically categorized by length, they can also be classified by chromosomal position (8). Long intergenic RNAs (lincRNAs) include enhancer RNAs (eRNAs) transcribed from distal enhancer regions, intronic lncRNAs, overlapping lncRNAs, sense lncRNAs, antisense lncRNAs, and bidirectional lncRNAs (9,10). lncRNAs control gene expression and signaling cascades at the epigenetic, transcriptional, and post-transcriptional levels by interacting with chromatin, proteins, and RNA targets (7). RNA polymerase type II (RNAP2) transcribes lncRNAs, which have comparable methylation patterns across the gene body and undergo post-transcriptional changes like splicing, polyadenylation, and 50 cappings (8). Despite lncRNAs being less stable than protein-coding genes, their promoter regions exhibit significant sequence stability, suggesting that lncRNA regulation is essential (7,10). LncRNA expression is tightly controlled and regulated at the epigenetic and transcriptional levels (11,12).
Despite their modest expression levels, lncRNAs exhibit a significant degree of tissue specificity and expression diversity (12).
Despite being strictly regulated in healthy tissues, lncRNAs are commonly dysregulated in illness, resulting in abnormal expressions and activities (13,14).
Recently, lncRNAs have been reported to be involved in cancer cell proliferation, metastasis, invasion, and apoptosis, with evidence that they serve as both tumor suppressors and oncogenes. LncRNAs are becoming more popular as possible anti-cancer therapy targets because of their involvement in critical pathophysiological processes (15). While some lncRNAs’ specific mechanisms of action have been fully described in LUSC, the vast majority of lncRNAs remain unknown (12).
Cancer immunotherapy is based on the idea of using the patient’s immune system to treat cancer (16). Cancer immune evasion is a complicated process that involves tumor gene mutation, tumor-associated antigens, a range of immune cells, and an inflammatory microenvironment (17). Tumor gene mutations can result in tumor neoantigens, rendering the immune system incapable of recognizing and eliminating tumor cells (18). Additionally, alterations in the tumor microenvironment might result in immune system dysfunction (19,20). Immunological checkpoints play a critical role in immune escape, and immunotherapy targeting immune checkpoints has emerged as a potential therapeutic option for patients with advanced NSCLC (21-23). Tremelimumab, an anti-cytotoxic T-lymphocyte-associated antigen 4 (anti-CTLA-4) drug, and nivolumab, an anti-programmed cell death protein 1 (anti-PD-1) agent, are now utilized in clinical practice to treat LUSC (24-26).
DNA methylation is a component of epigenetic changes that govern cellular activity and leads to cancer development (27). Lung cancer prognosis is influenced by epigenetic changes, especially DNA methylation (28-31). Novel prognostic and diagnostic markers in lung LUSC have been discovered using pharmacological suppression of DNA methylation together with gene expression (32). This review seeks to explore the clinicopathological impact of lncRNAs, immunotherapy, and DNA methylation in LUSC, emphasizing their exact roles in carcinogenesis and modes of action.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1607).
The role of lncRNA in cancer and mechanisms of action
Previously, genes were transcribed to mRNA and then translated to protein, with most of the genome being considered trash. Science and research have revealed that the once considered nonfunctional region of the genome contains RNAs that play many regulatory roles. The fast advancement in next-generation sequencing technologies has given rise to the concept of how genetic information gets translated from DNA to mRNA. This method resulted in the identification of several types of RNAs that were previously considered to be worthless (33).
RNAs are classified based on their function. Non-coding RNAs (ncRNAs) do not code for proteins, whereas coding RNAs (mRNAs) do. As the anticipated number of coding genes predicted to be discovered declines, thousands of non-coding genes (or RNAs) are found. The most well-known ncRNAs, rRNA and tRNA, are key players in protein synthesis and make up a significant component of the transcriptome. Non-coding RNAs are further classified into two groups depending on the number of nucleotides: those with fewer than 200 nucleotides (miRNA, siRNA, snRNA, and snoRNA) and those with 200 to 100,000 nucleotides (miRNA, siRNA, snRNA, and snoRNA) (34).
Classification of lncRNAs is a challenge due to their wide diversity of sizes, genome positions, and cell activities (35). LncRNAs are divided into two groups depending on their genomic location. Long intergenic non-coding RNAs, which are divided into antisense and sense, are the first kind. Long intronic non-coding RNAs are the second kind (36). LncRNAs, like mRNAs, can be spliced since they have a 5′ cap and a poly-A tail (37). Due to the nature of their promoters, lncRNAs are typically expressed by RNA polymerase II; however, they may be expressed by RNA polymerase III in specific conditions (38). Numerous illnesses have been implicated in lncRNAs, including neurological impairments, cardiovascular problems, and different types of cancer (36). While some lncRNAs are functionally defined by how genes up-regulate or downregulate, ribo-seq and mass spectrometry have revealed that certain lncRNAs include ORFs and are translated into functional proteins, despite their nomenclature. lncRNAs are classified according to six functional categories: (I) lncRNAs regulate protein expression to impact signaling pathways (39); (II) decoy lncRNAs function sponges for miRNAs, occupying decoy domains and inhibiting transcription factors (40); (III) scaffold lncRNAs serve as a foundation for the assembly and recruitment of substrates into polymeric complexes like RNPs (ribonucleoproteins), which are involved in ubiquitination and transcription (41); (IV) guide lncRNAs function as a scaffold for the assembly and recruitment of substrates to (42); (V) enhancer RNAs (eRNAs) enable chromatin looping so that enhancers and promoters may be connected (43), and (VI) lncRNAs containing ORFs, which may generate functional peptides (Figure 1) (9).
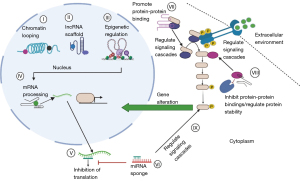
Our capacity to identify roles and categorize lncRNAs will improve as our knowledge of them develops. Epigenetics, mRNA and protein stability, splicing, and nuclear component trafficking are conceivable in biological processes. LncRNAs play a critical role in developing malignant cells, metastasis, and cancer progression through oncogenic and tumor-suppressive activities. LncRNAs can enhance oncogenic miRNA sponges and apoptosis and inhibit the cell cycle and the epidermal-mesenchymal transition (EMT), all of which have tumor-suppressing functions. Blocking the Wnt and PTEN pathways by downregulating MEG3 and up-regulating TP53, for example, may reduce certain cells development (44,45). MEG3 also functions as a miRNA sponge, preventing miR-421 and miR-21 from suppressing tumor growth (46). PANDAR, a lncRNA, may bind with BCL-2 and cause NSCLC cell death (47). PANDAR is reported in a range of malignancies as an oncogene (48). PANCR is another lncRNA that functions as a tumor suppressor. Its activity has an impact on NSCLC metastasis, presumably through regulating EMT. Patients who have a low PANCR level have a bad prognosis (49).
LncRNA dysregulation has recently been related to multi-drug resistance, cell proliferation, cell motility and invasion, apoptosis, metastasis, angiogenesis, and epithelial EMT. Colorectal cancer (CRC) (patients have a high level of lncRNA SNHG1 related to cell growth and progression (50). This RNA binds to PRC2, a histone modification enzyme that causes the downregulation of CDKN2B and KLF2. It also functions as a sponge for miR-154-5p, a CCND2 repressor. SNHG1 knockdown inhibits the progression of CRC cells. SiRNA downregulation of the lncRNA AFAP1-AS1 causes cell cycle arrest in the G2/M phase and apoptosis in esophageal cancer OE-33 cell lines (51,52). LINC00518 is a sponge for miR-199a and a multi-drug resistance lncRNA. MRP1 has been related to anti-cancer treatment resistance in various cancers, including breast cancer (BC), and miR-199a is reported to suppress it (53). MCM3AP-AS1 functions as a miR-211 sponge that causes angiogenesis-related proteins (AGGF1 and KLF5) to overexpress in glioblastoma cancer (54). MALAT1 promotes lung cancer metastasis via altering metastasis-related gene expression (55). The lncRNA ARNILA competes with SOX4 for miR-204 sponging in breast cancer, resulting in enhanced SOX4 expression and consequent metastasis and EMT (56). Because of their role in cancer, lncRNAs provide attractive therapeutic targets. Furthermore, their specificity for certain cancers and amounts in tumors and normal tissues make them effective diagnostic indicators; our understanding of such molecules is still evolving.
LncRNAs in lung cancer (LC)
Dysregulation of lncRNAs has been linked to a variety of illnesses, most notably cancer. Several studies have identified lncRNA expression patterns associated with lung cancer (LC) using next-generation sequencing methods (13,57). Wang et al. discovered 6,606 lncRNAs that were differently expressed between NSCLC and normal tissues. Another study discovered 856 differently expressed lncRNAs between LC and normal tissues by comparing four gene expression omnibus datasets. Previous research has demonstrated that numerous lncRNAs are dysregulated in lung cancer, implying that lncRNAs play a significant role in LC formation (58,59).
For instance, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) may facilitate LC metastasis to the brain via epithelial-mesenchymal transition since it was both highly expressed and linked with a poor outcome in NSCLC (60). Further study demonstrated that MALAT1 enhanced LC cell proliferation and resistance to gefitinib via sponging miR-200a, which controls ZEB1 expression in A549 cells (61). PVT1 expression was increased in NSCLC tissues, was linked with tumor stage and local metastasis, and maybe a target for therapy (62). HOTAIR expression was significantly increased in advanced-stage lung tumors. It was associated with metastases and a poor prognosis, and increased cancer cell migration and aggressiveness (63,64). ANRIL expression was also increased in NSCLC tumor tissues and related to tumor stage, size, and metastasis (65,66). DANCR expression was increased in LC, most notably in high-grade LC tissues and aggressive cancer cells. Ectopic expression of DANCR stimulated LC cell proliferation and colony formation, whereas silencing DANCR had the opposite impact (67). TUC338 expression was found to be much higher in LC than in noncancerous tissues. The survival period of TUC338 was shown to be associated with its expression. The examination of clinical data indicated a correlation between TUC338 expression and overall survival, tumor size, and lymph node metastasis in patients. Additionally, TUC338 may contribute to the development of LC via modulating the MAPK pathway (68). Collectively, these findings suggest that lncRNAs play a dysregulatory role in LC development.
lncRNAs in LUSC
Increasing evidence has demonstrated that lncRNAs play a role in the genesis of human cancer because of their regulatory influence on gene expression. Therefore, by discovering tumor-related lncRNAs and investigating their involvement in the onset and advancement of cancer, researchers may be able to find new therapeutic and diagnostic biomarkers for LUSC. Several lncRNAs are reported to be up-regulated (69-76), downregulated (77-79) in LUSC, and have tumorigenic functions in LUSC pathology. Comparable to protein-coding genes, lncRNAs can be classified as oncogenic or tumor-suppressive. This section discusses the role of lncRNAs as oncogenes and tumor suppressors in LUSC and their molecular pathways.
Oncogenic lncRNAs in LUSC
Tumors are caused and developed by dysregulation of gene expression, which often involves the activation of oncogenes or the inactivation of tumor suppressor genes. Oncogenes are genes whose products stimulate tumor growth, and oncogene dysregulation is a critical stage in cancer development. Numerous carcinogenic lncRNAs, including lncRNA-ATB, PITPNA-AS1, and LINC00173.v1, play critical regulatory roles in LUSC. As described in Table 1, these lncRNAs are increased in LUSC cells and enhance the development, proliferation, and invasion of LUSC cells.
Table 1
| LncRNA | Roles | Expression in cancer | Targeting approach | Effect in LUSC | Ref. |
|---|---|---|---|---|---|
| SNHG1 | Tumor suppressor | Up-regulated | TAp63/ZEB1 | Inhibit invasion, migration, and cell proliferation, promotes apoptosis | (69) |
| LINC000173.v1 | Oncogene | Upregulated | miR-511-5p/VEGFA | Enhanced migration, the proliferation of vascular endothelial cells and carcinogenesis | (70) |
| NNT-AS1 | Tumor suppressor | Up-regulated | miR-22/FOXM1 | Inhibits migration, and invasion, promotes cell death | (71) |
| LINC000519 | Tumor suppressor | Upregulated | miR-450b/miR-515-5p/YAP1 | Suppressed cell invasion and migration, induced cell death | (72) |
| LncRNA-ATB | Oncogene | Up-regulated | Inhibit miR-590-5p and activate the PI3K-Akt signaling pathway | Promotes migration, cell proliferation and invasion | (73) |
| MIR205HG | Oncogene | Upregulated | Bcl-2 and Bax/miR-299-3p/MAP3K2 | Enhanced migration, invasion, cell proliferation | (74) |
| LINC000355 | Tumor suppressor | Up-regulated | miR-466/LYAR axis | Inhibit cell proliferation, migration, and invasion, promoted cell apoptosis | (75) |
| LINC00466, DLX6-AS1 | Oncogene | Upregulated | miR-205 sponge/ CITED2 ENPP4 | Promotes metastasis, cell proliferation, inhibits apoptosis | (76) |
| MAGI2-AS3 | Tumor suppressor | Downregulated | miR-374a/b-5p/CAMD1 | Induced apoptosis, inhibits migration, invasion, and cell proliferation | (77) |
| PITPNA-AS1 | Oncogene | Downregulated | TAF15/ HMGB3 | Promotes migration, cell proliferation, and invasion | (78) |
| HULC | Oncogene | Downregulated | PTPRO/NFB | Enhanced cell proliferation | (79) |
| CASC9 | Oncogene | Up-regulated | – | Viability and proliferation | (80) |
In LUSC patients, upregulation of LINC00173.v1 was linked with a decreased overall and progression-free survival. LINC00173.v1 overexpression facilitated vascular endothelial migration and cell proliferation in vitro and in vivo, whereas LINC00173.v1 silencing reduced LUSC cell carcinogenesis. Additionally, LINC00173.v1 enhanced vascular endothelial migration, cell proliferation, and tumorigenesis in LUSC cells via sponging miR-511-5p. Notably, inhibiting LINC00173.v1 with the antisense oligonucleotide (ASO) approach inhibited LUSC cell tumor development and increased their therapeutic sensitivity to cisplatin in vivo. These findings elucidate the mechanism through which LINC00173.v1 stimulates the migration and proliferation of vascular endothelial cells and LUSC tumorigenesis, indicating that a LINC00173.v1-targeted medication in conjunction with cisplatin may be a suitable treatment for LUSC (70).
In another study, lncRNA-ATB overexpression enhances normal lung epithelial cell proliferation, migration, and invasive capacity.
Additionally, the migratory and invasive abilities of LUSC cells silenced by lncRNA-ATB are markedly reduced. Mechanistically, lncRNA-ATB binds directly to microRNA-590-5p and decreases its level while increasing NF-90 expression. Meanwhile, lncRNA-ATB overexpression promotes the epithelial-mesenchymal transition process, facilitating mesenchymal phenotype-related molecules vimentin and N-cadherin while inhibiting the expression of epithelial cells phenotype-related proteins E-cadherin and CK-19, in comparison to the control. MicroRNA-590-5p mimics, on the other hand, can counteract the effects of lncRNA-ATB overexpression. Altogether, these findings show that lncRNA-ATB overexpression may enhance LUSC development by altering the microRNA-590-5p/NF-90 axis. As a result, lncRNA-ATB may be prospective targets for LUSC treatment (73).
The levels of circulating noncoding RNA MIR205HG were significantly increased in LUSC tissues and cell lines (74,81). MIR205HG is needed for LUSC migration and cell differentiation. MIR205HG was reported to inhibit apoptosis in LUSC by regulating Bax and Bcl-2 (74).
Upregulation of PITPNA-AS1 stimulated the proliferation and migration of LUSC cells via TAF15-mediated stabilization of HMGB3. However, PITPNA-AS1 downregulation decreases migration and cell proliferation while increasing apoptotic activities of LUSC cells (78).
In LUSC tissue samples, lncRNA HULC was discovered to be downregulated. Additionally, lncRNA HULC expression was inversely correlated with protein tyrosine phosphatase receptor type O (PTPRO). Additionally, lncRNA HULC may enhance LUSC cell proliferation by inhibiting the production of PTPRO, a protein that is phosphorylated and activated by nuclear factor B (NFB). This study demonstrates the important function of lncRNA HULC in promoting LUSC by modulating the PTPRO/NFB signaling pathway, therefore identifying novel and potential LUSC biomarkers. However, the molecular mechanism by which lncRNA HULC lining interacts with PTPRO requires more investigation in a future study (79).
LncRNA CASC9 serves as an oncogene in LUSC and may be a promising target in LUSC diagnosis and prognosis. CASC9 knockdown by RNA interference may attenuate the viability and proliferation of LUSC cells. Using RT qPCR, Gao et al. compared CASC9 expression levels in LUSC and non-cancer tissues. LncRNA CASC9 was overexpressed in LUSC tissues compared to non-cancer tissues and significantly linked with malignant progression in LUSC. A statistical analysis of The Cancer Genome Atlas (TCGA) data and an integrated meta-analysis revealed higher CASC9 expression in LUSC tissues, thus validating the RT qPCR results (80).
Tumor-suppressive lncRNAs in LUSC
Tumor suppressor genes encode proteins that can prevent the growth of cancers. Mutations, deletions, or inactivation of tumor suppressor genes may result in cancer. Thus, a better knowledge of tumor suppressor genes may develop novel concepts for cancer-specific gene therapy. Tumor suppressor long non-coding RNAs are downregulated in LUSC, contributing to the disease’s genesis and progression. MAGI2-AS3, SNHG1, and MIR205HG are tumor suppressor lncRNAs, and we summarized their information in Table 1.
With downregulated expression, the cytoplasmic lncRNA MAGI2-AS3 was found to limit the proliferative, migratory, and invasive abilities of LUSC cells (77). MAGI2-AS3 overexpression also induced cell apoptosis. MAGI2-AS3 binds to miR-374a/b-5p, and through binding to CADM2 mRNA, it adversely modulates the mRNA and protein levels of CADM2. The inclusion of CAMD2 restores the biological activity-promoting effects of miR-374a/b-5p amplification. Finally, lncRNA MAGI2-AS3 inhibited LUSC via modulating the miR-374a/b-5p/CADM2 axis, suggesting that it might be used as a therapeutic biomarker for LUSC patients. However, this is only the beginning of MAGI2-AS3 research; other LUSC-related mechanisms need further investigation (77). LncRNA SNHG1 knockdown substantially reduces LUSC cell growth, metastasis, invasive ability, and triggered apoptosis. SNHG1 also influences zinc finger E-box binding homeobox 1 (ZEB1) expression, which is up-regulated in LUSC and promotes cell metastasis and invasion. Rather than interacting directly with ZEB1, SNHG1 regulates ZEB1 expression via inhibiting the function of the TAp63 isoform, a ZEB1 repressor that physically interacts with SNHG1. Furthermore, through the TAp63/ ZEB1 pathway, SNHG1 increases ZEB1 expression and boosts cell proliferation, metastasis, and invasiveness of LUSC cells while inhibiting apoptosis. These findings demonstrate that SNHG1 may have an oncogenic role in LUSC by suppressing TAp63 via the ZEB1 signaling pathway and that it may be a useful prognostic marker and treatment option for LUSC patients (69). LINC00519 was reported up-regulated in LUSC cells. A high LINC00519 level suggested a poor prognosis. In vitro, silencing LINC00519 inhibited proliferation, migration, and invasion and induced apoptosis in LUSC cells. Silencing LINC00519 in vivo reduced tumor development and lung metastasis. Mechanistically, LINC00519 was reported triggered via H3K27 acetylation (H3K27ac). Additionally, LINC00519 sponged miR-450b-5p and miR-51-5p to enhance the transcriptional regulator linked with Yes1 (YAP1). Moreover, miR-450b-5p and miR-51-5p induced anti-cancer activity in LUSC. Interestingly, miR-450b-5p and miR-51-5p expression levels significantly decreased in LUSC tissues and cells, suggesting that these miRNAs acted as suppressors in LUSC cells by inhibiting proliferation, migration and invasion, promoting apoptosis. Meanwhile, overexpression of YAP1 reversed the inhibitory effect of LINC00519 silencing on LUSC cell growth, indicating that LINC00519 was targeting YAP1 to control LUSC development. This study might help pave the way for new LUSC therapy options. However, additional factors affecting the pathway revealed in this study may require further examination (72). MIR205HG silencing significantly decreased cell proliferation, migration, and epithelial-to-mesenchymal transition (EMT) development, while increasing apoptosis. MIR205HG was shown to interact with miR-299-3p and silencing MIR205HG increased miR-299-3p expression. MAP3 K2 served as a miR-299-3p target gene and was up-regulated in response to MIR205HG overexpression. Overexpression of MAP3 K2 may function as a barrier against the impacts of MIR205HG downregulation on LUSC development. Thus, by targeting miR-299-3p in LUSC, MIR205HG functions as a ceRNA to promote cell proliferation and advancement (81).
LUSC has also been linked to the long non-coding RNA NNT-AS1. NNT-AS1, a novel lncRNA associated with cancer, has been identified as an oncogene implicated in the development of tumor cells in a variety of malignancies (82,83). The elevated expression of the lncRNA NNT-AS1 in LUSC tissues suggests that NNT-AS1 may have a regulatory function in LUSC. In LUSC, deletion of NNT-AS1 inhibited migration and invasion while inducing cell death. Additionally, the molecular investigation indicated the presence of an NNT-AS1/miR-22/FOXM1 regulatory network in LUSC and that suppressing NNT-AS1 had anti-tumor effects in LUSC via the miR-22/FOXM1 axis, shedding fresh light on the disease’s pathogenesis (71).
lncRNAs are also thought to have a role in oxidative stress regulation in LUSC. For example, LINC00355, primarily located in the cytoplasm, was elevated in LUSC and linked with a poor overall survival (OS) rate in LUSC patients. Furthermore, LINC0035 acted as a ceRNA for miR-466. LINC00355 expression was negatively correlated with the level of miR-466 and positively correlated with LYAR expression, a miR-466 target. By targeting miR-466, LINC00355 knockdown inhibited cell proliferation, migration, and invasion, promoted apoptosis in vitro and suppressed tumor development in vivo, therefore downregulating LYAR expression. These studies provide new insight into the molecular processes behind LUSC and suggest that LINC00355 may be utilized as a biomarker for LUSC diagnosis and therapy (75). Altogether, these findings demonstrate that lncRNAs have a tumor-suppressive function in LUSC. However, further work is required to assess tumor suppressor lncRNA delivery as a therapeutic option in the clinic. Table 1 illustrates some oncogenic and tumor-suppressive lncRNAs in LUSC.
lncRNAs in LUSC therapy
LUSC is the second most prevalent NSCLC and is often challenging to treat due to its frequent detection at a more advanced stage. Smoking has a strong connection to LUSC, which is frequently accompanied by other diseases (84). Because of these characteristics, LUSC has a dismal prognosis despite curative surgical resection and adjuvant therapy. The standard first-line treatment for LUSC is platinum-doublet chemotherapy, with second-line chemotherapy utilized in a small percentage of patients. Molecularly targeted anti-EGFR agents and anti-VEGFR2 antibodies have recently been authorized as second-line therapies (85).
LUSC patients, on the other hand, do not respond meaningfully to these medications due to a lack of significant driving mutations (86,87).
Increasing genetic knowledge about LUSC has resulted in many attractive molecular targets, including the FGFR gene (88), and therapeutic options to treat them. As a result, preclinical models are critical for validating molecularly targeted medicines and developing successful treatment methods such as combination therapies, including immunotherapy and DNA methylation. Chemotherapeutic sensitivity is still a significant challenge in LUSC therapy. Increasing evidence has revealed that lncRNAs induce carcinogenesis in different cancer types. However, knowledge of the biological roles and regulatory mechanisms of lncRNAs in response to cisplatin is not much. Li et al. discovered that the highly expressed lncRNA SFTA1P (surfactant associated 1, pseudogene) was down-regulated in LUSC tissues and may be activated by cisplatin therapy in LUSC cells. Increasing SFTA1P levels caused apoptosis in LUSC cells and made them more susceptible to cisplatin. Further experiment discovered that hnRNP-U, a ribonucleoprotein, was down-regulated in LUSC and linked to patients’ poor prognosis. SFTA1P might up-regulate hnRNP-U expression, according to mechanistic investigations. Furthermore, hnRNP-U increased cisplatin-induced apoptosis via up-regulating GADD45A, which high expression is linked to a favorable progression in LUSC patients. lncRNA SFTA1P may serve as a diagnostic for LUSC diagnosis and a predictor of cisplatin treatment response in LUSC patients (89).
In vivo, LINC00173.v1 increased the therapeutic sensitivity of LUSC cells to cisplatin, implying the possibility of LINC00173.v1 acting as a chemotherapeutic sensitizer in LUSC (70). LINC00173 expression was strongly associated with chemoresistance and progression in chemoresistant LUSC cell lines and patients. Additionally, LINC00173 enhanced β-catenin translocation by up-regulating the expression of Etk, GSKIP, and NDRG1 by sponging miRNA-218 as a ceRNA, resulting in the chemoresistance and development of LUSC tumors in vivo (90). As a result, LINC00173 has been identified as a possible therapeutic target in LUSC. The genes VEGFA, ATF4, and FN1, and the lncRNAs lncAP000769.1-2:10 and lncHFE2-2:1, are identified as having differential expression levels in celecoxib-treated human LUSC SK MES 1 cells compared to untreated cells. Additionally, these differential genes and lncRNAs have proven to have a high level of enrichment in pathways, including “protein processing in the endoplasmic reticulum”, “mTOR signaling pathway”, and “ECM receptor interaction”, among others. LncRNA AP000769.1-2:10 may modulate VEGFA gene expression in the mTOR signaling pathway, potentially enhancing celecoxib’s anti-tumor activity in LUSC therapy (90). However, further study is needed to explain the underlying molecular mechanisms involved with these lncRNAs and genes in LUSC.
lncRNAs as diagnostic and prognostic biomarkers in LUSC
LUSC is linked with a poor prognosis, primarily due to poor detection and late diagnosis, with a 5-year survival rate of about 15% (91). Traditional detection methods have limited sensitivity and specificity, making early detection difficult, and new biomarkers are needed for better molecular diagnosis and prognosis (92). Although proteins are widely employed as diagnostic markers, lncRNAs benefit from being stable and tissue-specific, and detectable in various physiological fluids (92,93). Patients may tolerate lncRNA-based biomarkers better than conventional biopsies since they are less intrusive (93). lncRNAs might be utilized to differentiate early-stage cancer patients from healthy controls with high sensitivity and specificity and provide prognostic information on the likelihood of metastasis and recurrence (94). For example, prostate cancer antigen 3 (PCA3) is an overexpressed lncRNA in prostate cancer (PCa) that contributes to PCa development by altering androgen signaling, and PCA3 urine levels have been effectively utilized as a PCa biomarker (95).
LINC00511 is overexpressed in LUSC tissues and cells, and its expression in LUSC patients is linked to TNM stages. LINC00511 affects LUSC progress by encouraging pro-migration and pro-proliferation, supporting the theory that LINC00511 is a potential oncogene implicated in LUSC development (96). Zhang et al. confirmed that LINC01133 was elevated in LUSC in an expression cohort of LUSC tissue and matching non-tumor tissues. Patients with increased LINC01133 expressions had lower survival periods, suggesting that LINC01133 might be used as a LUSC biomarker. Furthermore, in vitro investigations revealed that silencing LINC01133 decreased the ability of LUSC cells to invade, implying that LINC01133 may have a role in LUSC metastasis (97). The lncRNA FAM201A overexpression is linked with tumor progression in patients with advanced LUSC. Ectopic-inclined FAM201A significantly increased the proliferation and development of LUSC cells in vitro and in vivo. Additionally, FAM201A is a highly predictive biomarker of squamous cell carcinoma metastasis. Therefore, FAM201A may be a marker for distant metastases and poor survival in patients with LUSC (98).
Circulating ncRNAs can be used as diagnostic markers (99,100). In both LUSC clinical samples and cell lines, MIR205HG, a circulating lncRNA, was elevated. Interestingly, the amount of MIR205HG expression was positively linked with tumor size and differentiation from the clinical parameters of involved patients (74).
The rapid development of RNA-Seq analysis tools has given researchers a new viewpoint on the molecular characteristics and pathophysiology of LUSC and considerable evidence for predicting prognosis. Using several bioinformatics methods, Hu et al. explored LUSC-related RNA-seq data from The Cancer Genome Atlas (TCGA) database. They discovered that three lncRNA signatures in the ceRNA network (CACNA2D3- AS1, POU6F2-AS2, and TTTY16) were linked to overall survival (OS) in LUSC patients (101). CACNA2D3-AS1 was up-regulated in the ceRNA network, competing with has-mir-140, which was down-regulated. In many cancer cases, hsa-mir-140 acted as a tumor suppressor (102-104). Down-regulation of hsa-mir-140 promotes cell proliferation and invasion in various malignancies (102,103). Furthermore, hsa-mir-140 low expression is linked with a poor prognosis in spinal chordoma (105). Thus, abnormal CACNA2D3-AS1 expression may serve as a possible prognostic marker for LUSC. Similarly, POU6F2-AS2 was significantly expressed in LUSC tissue, and LUSC patients with high expression had low-risk scores. Meanwhile, TTTY16 in the ceRNA network was down-regulated and correlated with a good prognosis in LUSC (101).
Using a ceRNA network to analyze the genome-wide lncRNA expression patterns from TCGA, Sui et al. discovered a two-lncRNA signature PRR26 and FMO6P as a possible outcome predictor for LUSC patients. They discovered that LUSC patients with low-risk scores had a higher correlation with OS (106). Cheng et al. examined the diagnostic potential of lncRNAs in detecting early-stage LUSC. A panel of over four lncRNAs was identified and optimized using training and validation data from a separate independent cohort. Combining these lncRNAs gave much power in differentiating early-stage LUSC samples from non-tumor samples (107). Altogether, these findings imply that lncRNAs are promising diagnostic and prognostic markers in LUSC. However, regardless of the clinical relevance of these findings, certain conclusions are drawn using pure bioinformatics approaches on the TCGA database with no experimental examination. Therefore, further experimental research is necessary before these lncRNAs can be used in the clinic as diagnostic tools for LUSC. Table 2 illustrates some potential clinical applications of lncRNAs in LUSC
Table 2
| LncRNA | Expression in cancer | Link in LUSC | Ref |
|---|---|---|---|
| LINC00511 | Upregulated | TNM stages, promotes pro-proliferation, pro-migration | (96) |
| LINC01133 | Upregulated | Low survival periods, metastasis | (97) |
| FAM201A | Upregulated | Predictor of distance metastasis, and poor survival | (98) |
| CACNA2D3-AS1, POU6F2-AS2 | Upregulated | Prognostic biomarker in overall survival | (101) |
| TTTY16 | Downregulated | Prognostic biomarker in overall survival | (101) |
| lncRNA SOX2-OT | Up-regulated | tumor size, TNM stage, and lymph node metastasis | (108) |
| AL161431.1, LINC02389, APCDD1L-DT | – | LUSC prognosis | (109) |
| VPS9D1-AS1 | Up-regulated | Prognostic biomarker in overall survival | (110) |
| MALAT-1 | Upregulated | Prognostic biomarker in overall survival | (111) |
| LINC01031, LINC01088, LINC01931 | Upregulated | Poor survival | (112) |
| GATA6-AS1, TBX5-AS1, FEZF1-AS1, SFTA1P, LINC00968 | Up-regulated | Diagnosis and prognosis associated | (113,114) |
| FAM83A-AS1, MIR31HG, MIR99HG | – | Prognostic biomarker in overall survival | (115) |
| LINC00519 | Upregulated | Disease free survival | (116) |
Smoking-related LUSC diagnostic and prognostic biomarkers
Cigarette smoking has been linked to LUSC and can disrupt genetic characteristics directly or indirectly (108,109). Cigarette carcinogens may induce cancer by causing mutations in essential growth-regulating genes, including KRAS and TP53 (110). In addition, cigarette smoking leads to epigenomic dysregulation. First, smokers’ DNA methylation patterns differ from nonsmokers’, potentially disrupting genetic alterations in the small airway epithelium (111). Cigarette smoking also causes histone changes (112). Cigarette smoke causes site-specific post-translational histone modifications (PTMs) of histones H4 and H3, contributing to lung cancer development (113). Although cigarette usage is down worldwide as people become more aware of the dangers, lung cancer rates have not decreased. Hence, the identification of smoking-related LUSC diagnostic and prognostic biomarkers is paramount.
In smoking-related LUSC, biomarkers such as protein-coding genes, lncRNAs, and transcription factors have been reported. Recently, an eight-gene signature was created to help detect the survival risk in the LUSC dataset based on smoking status. Among these gene signatures, PYGB, WNT7A, and SLC7A5 were discovered with a greater probability of contributing to LUSC progression, suggesting that they might be utilized as smoking biomarkers. The epigenetic effect of smoking on cancer may be linked to single-copy deletion of SLC17A5 and WNT7A, as well as low-level amplification of PYGB. Endothelial cells, Activated NK cells, and dormant NK cells may all be involved in the unfavorable tumor microenvironment and immunologic dysfunction caused by cigarette smoking, resulting in tumor carcinogenesis, metastasis, and invasion (114). Additionally, Lung cancer survival has been related to the protein-coding genes AURKA and BIRC5 and the lncRNA LINC00094. The transcription of these genes and lncRNA might be necessary for LUSC prognosis and diagnosis in smokers with LUSC, which may help in understanding the disease’s pathogenesis (115).
Furthermore, carcinogens in cigarettes may cause cancer by affecting the host’s immune system (116,117). However, the Infiltration of particular immune cells, such as T follicular helper (Tfh) cells, has improved overall survival. Tfh cells are reported to show more significant infiltration in current smokers and LUSC current reformed smokers who have been smoking for less than 15 years, whereas resting memory CD4 T cells had reduced infiltration. Tfh cell infiltration has been linked to a higher overall survival (OS) rate; however, the relationship varied depending on the TNM stage (118).
Immunotherapy in lung cancer (LC)
Over the previous decade, several breakthroughs have been achieved in the field of cancer research. The discovery of innovative therapeutic agents and adopting an interdisciplinary strategy have revived optimism in people conquering cancer. However, lung cancer survival has not improved significantly at the population level. A rising proportion of lung cancer patients are being diagnosed early as low-dose computed tomography (CT) becomes more extensively embraced and employed. A detailed understanding of early-stage lung cancer and the finding of accurate prognostic-related genes are required for guiding appropriate therapy for patients with a poor prognosis. Given the human immune system’s critical involvement in the onset and development of LUSC (119-121), It will be highly beneficial to create a model for early-stage LUSC immune-based genes.
Immune checkpoint inhibitors (ICIs)
PD-L1 and PD-L2 are programmed death ligands that interact with PD-1 to inhibit T cells (Figure 2). PD-L2/PD-1 contact has a 26-fold greater affinity than PD-L1/PD-1 interaction (122). Additionally, PD-L1 can decrease T cell activity by attaching to the cluster of differentiation 80 (CD80) on activated T cells. In the other direction, PD-L2 can interact with the repulsive guidance molecule B (RGMB) receptor, a co-receptor for bone morphogenetic protein (BMP), activating T cells (123). Anti-PD-1 antibodies disrupt the PD-1/PD-L1 and PD-1/PD-L2 pathways but not the PD-L1/CD80 axis, inhibiting T cell activation. Additionally, anti-PD-1 indirectly increases the PD-L2/RGMB interaction, increasing the anti-tumor immune response mediated by T cells. Unlike PD-1 antibodies, PD-L1 antibodies also inhibit the PD-1/PD-L1 and PD-L1/CD80 axes but do not impact the PD-L2/PD-1 axis, resulting in tumor immune escape (124). This growing knowledge of the biochemical and signaling consequences will shed insight into the molecular processes behind PD-1 checkpoint blockade resistance and promote the establishment of combination treatments targeting PD-1 and its downstream targets.
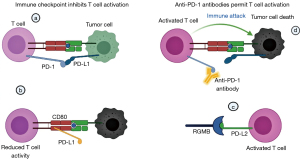
Immune checkpoint inhibitors stimulate effector T cells, which promote tumor blood vascular normalization. Normalizing tumor vessels increases the infiltration and activity of effector T cells, increasing anti-tumor immunity (125). This self-reinforcing feedback loop between immune reprogramming and tumor vascular normalization promotes immunological-mediated tumor elimination (126). The interplay between immune reprogramming and tumor vasculature justifies combining immunotherapy and antiangiogenic treatment
Effect of biomarkers in immunotherapy
Only a small percentage of patients have had long-term improvements from immune checkpoint inhibitors, and reliable biomarkers that may correctly identify these individuals before or early in the therapy process have yet to be discovered. Biomarkers including PD-L1, TMB, and tumor inflammation can help physicians figure out which patients will benefit the most from new drugs and combinations. While PD-L1 has numerous severe flaws as a predictive biomarker of long-term benefit, TMB is still being studied therapeutically.
In first-line treatment for PD-L1 expression of less than 50%, studies have indicated that ICIs increase OS relative to chemotherapy. Patients with squamous and non-squamous NSCLC, independent of PD-L1 expression level, benefit from a combination of ICIs and chemotherapy. However, a small percentage of PD-L1 TPS patients of less than 1% can benefit from immunotherapy alone (85), indicating that PD-L1 is an imprecise biomarker. IHC staining for PD-L1 in NSCLC is insufficient to detect prospective responders to PD-1/PD-L1 blockade-based immunotherapy (127,128). In addition, tumor heterogeneity, samples from primary versus metastatic lesions, various detection antibodies and cut-offs, staining methods, and immune escape from PD-L1/PD-1 inhibition are all important variables that may influence PD-L1’s predictive value (129-131). PET-based PD-L1 imaging using zirconium-89-labeled atezolizumab (132,133) is one way to overcome the spatial resolution restriction. The approach, which involves noninvasive imaging of tumor PD-L1 expression in vivo, might be used to help patients choose between anti-PD-1/PD-L1 therapy and track PD-L1 expression throughout treatment (132). The preliminary findings of first-in-human research to examine the practicality of imaging with zirconium-89-labeled atezolizumab and test its ability to predict a clinical response to PD-L1 inhibition in NSCLC were given by Bensch et al. (133). Tumor biopsy tissue gathered before, during, and after therapy will likely permit increased sophistication in treatment selection and immune elimination of cancers in the future.
Immunotherapy in LUSC
Immunotherapy has shown promise in NSCLC treatment, notably LUSC and adenocarcinoma. On the contrary, immunotherapy improves just a tiny portion of the patients’ population. Understanding the processes behind the varied immunotherapeutic responses is critical for enhancing personalized diagnosis and precision treatment. The anti-PD-1/PD-L1 therapy response is predicted by PD-L1 expression, a vital biomarker. CTLA-4 is vital for CD4+ T cell activity regulation and is implicated in immune response at the priming phase. The connection between the immunotherapeutic and other immunological biomarkers has been investigated to examine the immune signature’s prognostic value for LUSC immunotherapy. CTLA-4, CD274, and PDCD1 expression were much higher in low-risk patients than high-risk patients, confirming that matching antibodies may substantially affect low-risk individuals.
Additionally, patients with a low-risk score had much more tumor-infiltrating cells and cytotoxic activity than those with a high-risk score. As a result, immunotherapy may benefit low-risk individuals more than high-risk ones (134). However, there are significant drawbacks to this study. First, data for evaluating immunotherapy were extracted from the TCGA dataset rather than from a genuine immunotherapy cohort. Second, the current study used a limited sample size, and bigger sample size is required to enhance the reliability of the immunological signature. Thirdly, functional investigations using immunological signatures are needed to deduce the molecular process. As a result, further study is necessary to resolve these difficulties.
Given the well-established involvement of inflammatory cytokines in the carcinogenesis, aggressiveness, and metastasis of LUSC, these cytokines may be utilized as clinical markers to monitor disease progression or therapeutic targets (135,136). Additionally, most genes linked to the immune system have undergone genetic alterations such as amplification and profound deletion, correlated with clinical outcomes (137). A recent study indicated that cancer immune invading cells are closely related to clinical outcomes (138,139). Recent research examined the link between risk signatures and immune invading cells. Notably, the risk signature score was positively linked with the infiltration of CD8+ T cells, neutrophils, dendritic cells, and macrophages, implying that the levels of these four immune cells may be increased in high-risk individuals and the signature may predict immune cell infiltration (140). An earlier study discovered that early proliferative CD8 T+ cell responses are linked to an improved prognosis in patients treated with a PD-1 inhibitor (141). Macrophages make up most tumor-immune infiltrating cells, and their polarization determines their oncogenesis consequences (142). Several studies have revealed a clear correlation between macrophage quantity, phenotype, and survival in individuals with LUSC (143,144). In general, this novel signature can be utilized as a factor that predicts LUSC and LUSC patients’ immunological status.
Given the complex genetic structure of LUSC, it is a heterogeneous disease with no effective treatments. Recently, it was established that tumor immune response plays a crucial role in LUSC genesis and development (145,146). According to certain research, an immunogenomic or immune infiltrating cell-based risk profile for NSCLC, particularly LUSC and adenocarcinoma, may be utilized as a stand-alone prognostic factor (147-149). Apelin (APLN), a ligand for the G protein-coupled receptor, is an angiogenic factor contributing to tumor angiogenesis (150). Previous studies reported that increased APLN expression is related to a poorer OS rate in NSCLC patients (151,152). Fibroblast growth factor receptor 4 (FGFR4) is linked with oncogenesis, is prognostic in LUSC, and plays a critical role in the tumor microenvironment (153-155). Semaphorin 4C (SEMA4C) controls immune cell interactions, angiogenesis, and tumor formation (156). SEMA4C deficiency inhibits NSCLC cells development and reverses the epithelial-mesenchymal transition (157). Hou et al. developed a predictive immunogenomic marker for LUSC and proved its usefulness in predicting clinical outcomes in LUSC. The findings revealed that APLN expression was substantially higher in LUSC patients, although FGFR4 and SEMA4C expression were higher in male LUSC patients without metastases (140). When these three genes are considered in conjunction with past studies, they may represent LUSC risk factors.
Immune microenvironment and tumor mutations both contribute significantly to LUSC prognosis and progression. Recent research developed and tested an immunological profile based on five genes (FLNC, RND1, IL4I1, TGM2, and PTGIS). The signature was capable of predicting LUSC patients’ prognosis and immunotherapy response. At each tumor stage of the immunological signature, patients with a high-risk score had a considerably poorer prognosis than patients with a low-risk score. Numerous Gene Expression Omnibus (GEO) datasets indicated that the nomogram that combined the immunological signature with the tumor stage accurately predicted LUSC patient survival. Additionally, patients with a low-risk score may have a better chance of responding to immunotherapy (134). In another study, a risk model based on 17 immune-related genes was developed and tested to generate a prognostic prediction of early-stage LUSC patients. The mechanisms by which this model predicts LUSC prognosis in the early stages was clarified by identifying gene mutation profiles and immune cell infiltration characteristics, which may aid clinicians in implementing effective lung cancer treatments for patients, particularly individualized treatment for those patients with specific gene mutations and tumor immunotherapy (158).
DNA methylation in LUSC
Despite significant advancements in the prevention, diagnosis, and treatment of LUSC in recent decades, the 5-year survival rate remains poor, at under 15% (159). With the rise in poor prognosis and advanced diagnosis in LUSC recently, it has become more important to investigate new effective prognostic and diagnostic indicators for predicting LUSC survival. According to growing data from epigenetic studies, DNA methylation is important in gene expression and occurs early in some cancers. DNA methylation is one of the first observable neoplastic alterations, giving it a distinct advantage as a biomarker for cancer prognosis and diagnosis (160-162). Furthermore,
methylation is a significant epigenetic alteration of genomic DNA and a critical mechanism of genomic function regulation. Although epigenetic changes are reversible, they have tremendous therapeutic promise (163). Moreover, the use of RNA-Seq data processing and DNA methylation to uncover disease-specific prognostic and diagnostic biomarkers in lung cancer offers a unique approach (164,165). As a result, detecting and treating DNA methylation as a target will develop novel lung cancer diagnostics and therapy methods.
Using methylation data from patients with LUSC, Li et al. developed a LUSC prognosis model (166). Examination of the ROC curve multi-year survival rate curve reveals that the model’s prediction results improve in line with survival time growth. The developed prediction model incorporated independent prognostic methylation sites and may be utilized to discover novel tumor indicators, guide therapeutic therapy, and assess patient prognosis. Additionally, by categorizing the genetic subtypes of LUSC patients, this model can give more tailored therapy recommendations and prognosis assessments. (166). Zhang et al. developed the first risk prognosis model for LUSC patients based on DNA methylation sites, demonstrating improved stability and reliability and the best predictor of OS for LUSC patients. Additionally, to facilitate clinical decision-making using the risk model, they constructed a nonogram incorporating the DNA methylation signature, tobacco smoking history, and distant metastatic stage (167). In another study, Zhang et al. discovered that methylation of the TRIM58/cg26157385 locus was related to the expression of eight prognostic genes in LUSC patients (168). However, before the approach can be applied in the clinic, more clinical studies are required to determine the robustness of this DNA methylation pattern. During the process of choosing prognostic-related DNA methylation sites for a model, certain deviations are unavoidable. Correlation analysis indicates that future research should combine mRNA and DNA methylation signatures to develop a more accurate predictive biomarker (168). Wang et al. discovered that in LUSC tissue, AKAP13 mRNA expression was decreased while methylation was increased. Three AKAP13 CpG sites were found to be linked to OS. AKAP13 mRNA and methylation analysis identified 11 CpG sites linked with OS in LUSC patients. Although the expression of AKAP13 mRNA was related to distant metastasis in LUSC, no correlation was seen between the methylation status of CpG sites and clinical characteristics. Therefore, AKAP13 mRNA and its methylation CpG sites could be utilized as prognostic markers in individuals with LUSC (169).
Previous studies established that hypomethylation-induced increased gene expression and hypermethylation-induced reduced gene expression play critical roles in lung cancer regulation and development (170,171). For example, Sugimoto et al. discovered that patients benefit from aberrant GRWD1 (glutamate-rich WD repeat-containing 1) methylation during tumor development, as the GRWD1 gene’s activity is inhibited in tumor cells by its methylation.
In contrast, GRWD1 gene expression promotes tumor cell growth (172). By contrast, Chen et al. discovered that hypermethylation of the AGTR1 promoter is more prevalent in LUSC patients (173). Ni et al. have shown that the SHOX2 (short stature homeobox 1) gene methylation is more prominent in lung cancer, particularly LUSC, and may serve as a noninvasive lung cancer diagnostic (174). Guo et al. discovered that the hypermethylated status of the WIF-1 gene, which is frequently observed in NSCLC, is more common in squamous cell carcinomas and correlates with a poor clinical prognosis (175). Kim et al. discovered that excessive methylation of the Wrap53 promoter predicts a poor outcome in individuals with borderline significance in a research using tumor and matching non-malignant lung tissue specimens (176). According to Zhang et al., hypermethylation of the PAX6 gene is an independent prognostic indicator and is substantially associated with a low OS rate in NSCLC, making it a potentially promising biomarker for prognostic evaluation in NSCLC patients (177). According to an integrated survival, methylation, and gene expression study, ZNF418 indicates a poor prognosis in LUSC patients (178). In LUSC, the G protein-coupled receptor GPR75 is hypermethylated, which is linked to LUSC failure. STX12 falls in the syntaxin family of SNAREs protein receptors. It has been discovered that STX12 expression is inversely connected with methylation, and STX12 hypermethylation is linked to better clinical outcomes in LUSC (178). Rui et al. used MethylMix to find methylation-driven genes in LUSC tumors and normal samples from the TCGA database to predict LUSC prognosis. Cox regression studies on multivariate and univariate data revealed that twelve abnormally methylated genes in a Cox prediction model were related to OS in LUSC patients. The combined survival study of methylation and gene expression revealed that WDR61 and DQX1 had a low survival rate due to hypermethylation and low expression. DQX1 expression was inversely associated with the methylation site cg02034222. Thus, WDR61 and DQX1 may serve as promising biomarkers for determining LUSC prognosis (179). In another research, three hub methylated-driven genes (MDGs), ZNF454, PMPCAP1, and SOWAHC, were discovered to have independent prognostic values in LUSC (180). Using the MethylMix algorithm, PMPCAP1 and SOWAHC were discovered to be hypomethylated and overexpressed in individuals with LUSC, indicating a bad prognosis. By contrast, ZNF454 was hypermethylated and expressed at a reduced level, predicting an improved prognosis. Enrichment analysis showed that ZNF454, PMPCAP1, and SOWAHC were predominantly engaged in transcription or gene expression pathways. Thus, the Methylation-driven genes (MDGs) ZNF454, PMPCAP1, and SOWAHC may serve as predictive biomarkers for LUSC, guiding diagnostic and treatment choices and creating a theoretical foundation for future research (180). Collectively, a thorough survival and functional study of methylation-driven genes enables a better knowledge of their underlying processes and the identification of innovative lung cancer therapeutic options. Table 3 illustrates some DNA methylated genes and their roles in LUSC.
Table 3
| DNA methylated gene | Role in LUSC | Ref. |
|---|---|---|
| FZD10, FZD2, GNAS | LUSC prognosis | (166) |
| RASSF6, ZNF773, HES7, SPC25, TRIM71, LHX5, APOBEC3C, RPS18, ISL2, INSM2 | LUSC prognosis | (167) |
| A2ML1, CCNE1, COBL, ESCO2, GPR115, MMP10, OVOL1, SCGB1A1 | LUSC prognosis | (168) |
| AGGF1P3, HIST1H4K, CH17-140K24.1, ADCYAP1, COX11P1, FOXD4L6, CBLN1, ATP6V0CP3, RP11-264L1.4, LINC01158, CTC-523E23.14, TRIM58 | Overall survival | (179) |
| ZNF454, PMPCAP1, SOWAHC | LUSC prognosis | (180) |
| GCSAM, GPR75, NHLRC1, TRIM58 | LUSC prognosis | (181) |
Limitations and future prospectives
Despite data supporting the therapeutic significance of lncRNAs in LUSC, there remain constraints to targeting and evaluating the efficacy of lncRNAs. For example, the absence of a protein product limits therapy options to mostly nucleic acid-based treatments (181). While they are effective, they have some drawbacks, including off-target effects, difficulties penetrating the cellular plasma membrane, and decreased bioavailability (182). Additionally, unlike proteins, the three-dimensional (3D) structure of lncRNAs is largely unknown, and a lack of conserved regions may make small-molecule inhibitor creation more difficult. A lack of complete understanding of the processes and regulatory networks governing many lncRNAs in LUSC may further hinder the development of particular targeted methods for toxicity reduction (181). While certain lncRNAs in LUSC are conserved across species, many are not, obstructing research and preclinical studies in animal models (181,182). Additionally, the lungs are challenging for siRNA and ASO delivery in vivo, which may impede preclinical investigations for lncRNA treatments in LUSC. Initial lung delivery is challenging due to physical obstacles such as mucosa and cilia and the need to evade the immune system. Additionally, passage across the target cell membrane into the appropriate subcellular compartment and evasion of endosome destruction is difficult to accomplish (183).
The overall response rate and the incidence of acquired resistance among PD-1 blockade responders appear to have an inverse relationship across tumor types (184). Enhancing antigenicity, modulating TME, boosting immune cell activity, and overcoming resistance mediated by other elevated immunological checkpoints are all strategies for combating immunotherapy resistance. Immune checkpoint targeted treatments (ICT) require mechanism-based methods to overcome resistance. Ishizuka et al. demonstrated that silencing the RNA-editing enzyme ADAR1 overcomes resistance to PD-1 checkpoint inhibition induced by antigen presentation inactivation in tumor cells (185). BMP7 suppresses proinflammatory responses in the TME by inhibiting mitogen-activated protein kinase 14 expression in macrophages and CD4+ T cells; according to Cortez et al. BMP7 knockdown combined with anti-PD1 stimulates CD4+ and CD8+ T cells in tumors, reduces M2 macrophages and resensitizes immunotherapy-resistant malignancies (186). According to new data, targeting epigenetic elements that promote tumor development and restrict immune cell function can improve anti-tumor immunity by altering the TME. Inhibition of the enhancer of zeste homolog 2, the catalytic subunit of polycomb repressive complex 2, can increase T regulatory cell trafficking, impair T regulatory cell capacity, improve antigen presentation, and increase anti-tumor immunity, making it a promising target for overcoming ICB resistance in some cancers (187).
The discovery of other targeted therapies for advanced LUSC is necessary to provide significant survival improvements and enable customized care for patients. Combinations of immunotherapy drugs and immunotherapies with targeted treatments form a multifaceted strategy for treating advanced LUSC. These methods are under investigation and may allow higher survival results.
Also, DNA methylation and RNA-Seq data analysis offer an innovative approach for identifying disease-specific diagnostic and prognostic biomarkers in cancer therapy (164,165). The fast advancement of RNA-Seq analysis methods enables a fresh viewpoint on the molecular characteristics and pathophysiology of LUSC and important evidence for prognosis prediction.
The epigenetic alteration, particularly DNA methylation, has a role in LUSC pathogenesis. Accumulating data indicates that DNA methylation is the primary biological process behind the epigenetic change in human malignant cancers, including lung cancer (188-190). Methylation alterations in a subset of tumor-associated genes have been discovered in prior tumorigenesis investigations, suggesting that they are significant risk factors for tumorigenesis and molecular markers for early detection (191). Because PR/SET domain 5 promoter methylation is strongly linked with lymph node metastases and tumor differentiation status in LUSC, this gene is a prospective target for the diagnosis, prognosis, and therapy of this malignancy (192). The drought-repressed 4 gene has been proposed for analyzing the methylation state of LUSC cells, and its low expression corresponds with a bad prognosis in patients (193). Methylation of the tripartite motif 58/cg26157385 is linked with the expression of eight prognosis-associated genes in LUSC, indicating a possible regulatory function in the development of LUSC (168). Thus, gene expression and bioinformatics analysis of DNA methylation pave the way for discovering disease-specific diagnostic and prognostic biomarkers in LUSC.
Conclusions
LncRNAs have been discovered to participate in a wide range of critical cellular functions, with their involvement in cancer becoming increasingly clear (7). Localization of lncRNAs is primarily determined by their function; they may interact with chromatin, protein, and RNA to control all stages of gene expression and affect critical signaling cascades (7). Numerous small non-coding RNAs were reported up-regulated and downregulated and act as oncogenes, promoting LUSC proliferation, survival, invasion, migration, metastasis, and EMT. Certain lncRNAs, such as MAGI2-AS3, SNHG1, NNT-AS1, and MIR205HG, have participated in many facets of LUSC carcinogenesis via several distinct pathways. While miRNA sponging seems to be the most often observed method of lncRNA regulation in LUSC, numerous lncRNAs can also regulate gene expression, protein interactions, and stability, indicating that lncRNA regulation in LUSC is complicated. Tumor suppressor lncRNAs that are downregulated in LUSC function similarly to oncogenes, although to inhibit cancer progression. Many of the pathways and genes controlled by suppressor lncRNAs are also regulated by oncogenic lncRNAs, therefore, carcinogenic lncRNAs may be increased to counteract the suppressor lncRNAs. Moreover, lncRNAs appear to have a role in the cisplatin-based chemotherapeutic response and may serve as indicators for treatment response and potential therapeutic targets in LUSC. While the mechanisms of action for most lncRNAs are discussed in length in this study, the means of action for many lncRNAs remain unknown and must be explained to determine their function as tumor promoters or suppressors. Given the poor prognosis linked to LUSC, primarily due to late diagnosis and a lack of effective therapies for late-stage illness, more innovative methods to LUSC care are necessary. High-throughput data mining establishes an efficient bioinformatics foundation for furthering our knowledge of the etiology and prognosis of LUSC, which has significant theoretical implications for future LUSC research.
The introduction of PD-1/PD-L1/CTLA-4-targeted treatment has changed LUSC patient management, resulting in long-lasting responses and improved OS. Immunotherapy, on the other hand, does not help all patients. Patients with primary or acquired resistance do not experience tumor shrinkage or more prolonged survival. Predictive biomarkers including PD-L1, tumor mutational burden (TMB), and tumor inflammation should be investigated further. Increased knowledge of biology and molecular subgroups of NSCLC has resulted in developing novel biomarker-directed treatments and improved OS in patients with advanced or metastatic illnesses. Patients lacking molecular therapeutic targets have been treated with immune checkpoint treatments in the first-line situation since the emergence of PD-1/PD-L1 blockers in 2015, notably for squamous NSCLC patients (194-196).
Meanwhile, modification of epigenetic markers, particularly DNA methylation, has a role in the pathophysiology of LUSC. Accumulating data indicated that DNA methylation is the primary biological process behind the epigenetic change in human malignant cancers, including lung cancer (188-190). The fast advancement of RNA-Seq analysis methods enables a fresh viewpoint on the molecular characteristics and pathophysiology of LUSC and substantial evidence for prognosis prediction in LUSC. However, the precise role of epigenetic aberrations and DNA methylation alterations in LUSC genesis is not fully known. Additional mechanistic investigations, including animal models of epigenomic dysregulation, are necessary. Early lung cancer detection is critical for the administration of curative treatment regimens, including surgery. Methylation biomarkers can become very sensitive diagnostic tools for early illness detection in blood serum or sputum. Although numerous potential methylation indicators have been identified, developing sensitive and accurate approaches for identifying uncommon modified DNA molecules in sputum, serum, or circulating tumor cells should now take precedence.
Acknowledgments
Funding: This work was supported by the Foundation of Science Technology Department of Zhejiang Province, China social development projects (LGF21C050001)
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1607
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-1607
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1607). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet 2011;378:1727-40. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Chansky K, Sculier JP, Crowley JJ, et al. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol 2009;4:792-801. [Crossref] [PubMed]
- Tsai JH, Donaher JL, Murphy DA, et al. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 2012;22:725-36. [Crossref] [PubMed]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018;18:5-18. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Mongelli A, Martelli F, Farsetti A, et al. The Dark That Matters: Long Non-coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front Physiol 2019;10:369. [Crossref] [PubMed]
- Enfield KS, Pikor LA, Martinez VD, et al. Mechanistic Roles of Noncoding RNAs in Lung Cancer Biology and Their Clinical Implications. Genet Res Int 2012;2012:737416. [Crossref] [PubMed]
- Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics 2016;14:42-54. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Roth A, Diederichs S. Long Noncoding RNAs in Lung Cancer. Curr Top Microbiol Immunol 2016;394:57-110. [Crossref] [PubMed]
- Wang L, Chen Z, An L, et al. Analysis of Long Non-Coding RNA Expression Profiles in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2016;38:2389-400. [Crossref] [PubMed]
- Zhan Y, Zang H, Feng J, et al. Long non-coding RNAs associated with non-small cell lung cancer. Oncotarget 2017;8:69174-84. [Crossref] [PubMed]
- Zhang Y, Tang L. The Application of lncRNAs in Cancer Treatment and Diagnosis. Recent Pat Anticancer Drug Discov 2018;13:292-301. [Crossref] [PubMed]
- Wei SC, Duffy CR, Allison JP. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov 2018;8:1069-86. [Crossref] [PubMed]
- Anichini A, Perotti VE, Sgambelluri F, et al. Immune Escape Mechanisms in Non Small Cell Lung Cancer. Cancers (Basel) 2020;12:3605. [Crossref] [PubMed]
- Anagnostou V, Smith KN, Forde PM, et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:264-76. [Crossref] [PubMed]
- Schoenhals JE, Seyedin SN, Anderson C, et al. Uncovering the immune tumor microenvironment in non-small cell lung cancer to understand response rates to checkpoint blockade and radiation. Transl Lung Cancer Res 2017;6:148-58. [Crossref] [PubMed]
- Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 2020;30:507-19. [Crossref] [PubMed]
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. [Crossref] [PubMed]
- Vansteenkiste J, Wauters E, Reymen B, et al. Current status of immune checkpoint inhibition in early-stage NSCLC. Ann Oncol 2019;30:1244-53. [Crossref] [PubMed]
- Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275-87. [Crossref] [PubMed]
- Ferrara R, Imbimbo M, Malouf R, et al. Single or combined immune checkpoint inhibitors compared to first-line platinum-based chemotherapy with or without bevacizumab for people with advanced non-small cell lung cancer. Cochrane Database Syst Rev 2021;4:CD013257. [PubMed]
- Hussaini S, Chehade R, Boldt RG, et al. Association between immune-related side effects and efficacy and benefit of immune checkpoint inhibitors - A systematic review and meta-analysis. Cancer Treat Rev 2021;92:102134. [Crossref] [PubMed]
- Hu-Lieskovan S, Lisberg A, Zaretsky JM, et al. Tumor Characteristics Associated with Benefit from Pembrolizumab in Advanced Non-Small Cell Lung Cancer. Clin Cancer Res 2019;25:5061-8. [Crossref] [PubMed]
- Cai Y, Tsai HC, Yen RC, et al. Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells. Genome Res 2017;27:533-44. [Crossref] [PubMed]
- Altenberger C, Heller G, Ziegler B, et al. SPAG6 and L1TD1 are transcriptionally regulated by DNA methylation in non-small cell lung cancers. Mol Cancer 2017;16:1. [Crossref] [PubMed]
- Grasse S, Lienhard M, Frese S, et al. Epigenomic profiling of non-small cell lung cancer xenografts uncover LRP12 DNA methylation as predictive biomarker for carboplatin resistance. Genome Med 2018;10:55. [Crossref] [PubMed]
- Kettunen E, Hernandez-Vargas H, Cros MP, et al. Asbestos-associated genome-wide DNA methylation changes in lung cancer. Int J Cancer 2017;141:2014-29. [Crossref] [PubMed]
- Nikolaidis G, Raji OY, Markopoulou S, et al. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res 2012;72:5692-701. [Crossref] [PubMed]
- Zhong S, Fields CR, Su N, et al. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene 2007;26:2621-34. [Crossref] [PubMed]
- Sousa-Franco A, Rebelo K, da Rocha ST, et al. LncRNAs regulating stemness in aging. Aging Cell 2019;18:e12870. [Crossref] [PubMed]
- Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol 2016;937:3-17. [Crossref] [PubMed]
- Sana J, Faltejskova P, Svoboda M, et al. Novel classes of non-coding RNAs and cancer. J Transl Med 2012;10:103. [Crossref] [PubMed]
- Dahariya S, Paddibhatla I, Kumar S, et al. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol 2019;112:82-92. [Crossref] [PubMed]
- Krchnáková Z, Thakur PK, Krausová M, et al. Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5' splice-site sequences due to weak interactions with SR proteins. Nucleic Acids Res 2019;47:911-28. [Crossref] [PubMed]
- Da Sacco L, Baldassarre A, Masotti A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int J Mol Sci 2012;13:97-114. [Crossref] [PubMed]
- Yang M, Wang M, Li X, et al. The role of lncRNAs in signaling pathway implicated in CC. J Cell Biochem 2019;120:2703-12. [Crossref] [PubMed]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol 2016;1402:271-86. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013;4:2939. [Crossref] [PubMed]
- Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res 2018;3:108-17. [Crossref] [PubMed]
- Liu Y, Ding M, Gao Q, et al. Current Advances on the Important Roles of Enhancer RNAs in Gene Regulation and Cancer. Biomed Res Int 2018;2018:2405351. [Crossref] [PubMed]
- Inamura K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs: Clinical Relevance in Lung Cancer. Cells 2017;6:12. [Crossref] [PubMed]
- Zheng Q, Lin Z, Xu J, et al. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting β-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis 2018;9:253. [Crossref] [PubMed]
- Youness RA, Gad MZ. Long non-coding RNAs: Functional regulatory players in breast cancer. Noncoding RNA Res 2019;4:36-44. [Crossref] [PubMed]
- Han L, Zhang EB, Yin DD, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis 2015;6:e1665. [Crossref] [PubMed]
- Li J, Li Z, Zheng W, et al. PANDAR: a pivotal cancer-related long non-coding RNA in human cancers. Mol Biosyst 2017;13:2195-201. [Crossref] [PubMed]
- Sun M, Liu XH, Wang KM, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 2014;13:68. [Crossref] [PubMed]
- Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer 2018;17:141. [Crossref] [PubMed]
- Rossi MN, Antonangeli F. LncRNAs: New Players in Apoptosis Control. Int J Cell Biol 2014;2014:473857. [Crossref] [PubMed]
- Wu W, Bhagat TD, Yang X, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology 2013;144:956-966.e4. [Crossref] [PubMed]
- Chang L, Hu Z, Zhou Z, et al. Linc00518 Contributes to Multidrug Resistance Through Regulating the MiR-199a/MRP1 Axis in Breast Cancer. Cell Physiol Biochem 2018;48:16-28. [Crossref] [PubMed]
- Yang C, Zheng J, Xue Y, et al. The Effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 Axis Regulating Glioblastoma Angiogenesis. Front Mol Neurosci 2018;10:437. [Crossref] [PubMed]
- Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73:1180-9. [Crossref] [PubMed]
- Yang F, Shen Y, Zhang W, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ 2018;25:2209-20. [Crossref] [PubMed]
- Qiu M, Feng D, Zhang H, et al. Comprehensive analysis of lncRNA expression profiles and identification of functional lncRNAs in lung adenocarcinoma. Oncotarget 2016;7:16012-22. [Crossref] [PubMed]
- Chen J, Wang R, Zhang K, et al. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med 2014;18:2425-36. [Crossref] [PubMed]
- Xu YJ, Du Y, Fan Y. Long noncoding RNAs in lung cancer: what we know in 2015. Clin Transl Oncol 2016;18:660-5. [Crossref] [PubMed]
- Shen L, Chen L, Wang Y, et al. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol 2015;121:101-8. [Crossref] [PubMed]
- Feng C, Zhao Y, Li Y, et al. LncRNA MALAT1 Promotes Lung Cancer Proliferation and Gefitinib Resistance by Acting as a miR-200a Sponge. Arch Bronconeumol 2019;55:627-33. (Engl Ed). [PubMed]
- Yang YR, Zang SZ, Zhong CL, et al. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol 2014;7:6929-35. [PubMed]
- Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun 2013;436:319-24. [Crossref] [PubMed]
- Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013;13:464. [Crossref] [PubMed]
- Lu Y, Zhou X, Xu L, et al. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther 2016;9:3077-84. [PubMed]
- Lin L, Gu ZT, Chen WH, et al. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol 2015;10:14. [Crossref] [PubMed]
- Zhen Q, Gao LN, Wang RF, et al. LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a. Cancer Control 2018;25:1073274818769849. [Crossref] [PubMed]
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci 2018;22:443-9. [PubMed]
- Zhang HY, Yang W, Zheng FS, et al. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in Lung squamous cell carcinoma. Biomed Pharmacother 2017;90:650-8. [Crossref] [PubMed]
- Chen J, Liu A, Wang Z, et al. LINC00173.v1 promotes angiogenesis and progression of lung squamous cell carcinoma by sponging miR-511-5p to regulate VEGFA expression. Mol Cancer 2020;19:98. [Crossref] [PubMed]
- Ma J, Qi G, Li L. LncRNA NNT-AS1 promotes lung squamous cell carcinoma progression by regulating the miR-22/FOXM1 axis. Cell Mol Biol Lett 2020;25:34. [Crossref] [PubMed]
- Ye P, Lv X, Aizemaiti R, et al. H3K27ac-activated LINC00519 promotes lung squamous cell carcinoma progression by targeting miR-450b-5p/miR-515-5p/YAP1 axis. Cell Prolif 2020;53:e12797. [Crossref] [PubMed]
- Li J, Xia R, Liu T, et al. LncRNA-ATB Promotes Lung Squamous Carcinoma Cell Proliferation, Migration, and Invasion by Targeting microRNA-590-5p/NF90 Axis. DNA Cell Biol 2020;39:459-73. [Crossref] [PubMed]
- Chang Y, Xue X, Li C, et al. MIR205HG facilitates carcinogenesis of lung squamous cell carcinoma in vitro revealed by long noncoding RNA profiling. Acta Biochim Biophys Sin (Shanghai) 2020;52:371-81. [Crossref] [PubMed]
- Sun X, Wang G, Ding P, et al. LINC00355 promoted the progression of lung squamous cell carcinoma through regulating the miR-466/LYAR axis. Braz J Med Biol Res 2020;53:e9317. [Crossref] [PubMed]
- Yao Y, Zhang T, Qi L, et al. Competitive Endogenous RNA Network Construction and Comparison of Lung Squamous Cell Carcinoma in Smokers and Nonsmokers. Dis Markers 2019;2019:5292787. [Crossref] [PubMed]
- He J, Zhou X, Li L, et al. Long Noncoding MAGI2-AS3 Suppresses Several Cellular Processes of Lung Squamous Cell Carcinoma Cells by Regulating miR-374a/b-5p/CADM2 Axis. Cancer Manag Res 2020;12:289-302. [Crossref] [PubMed]
- Ren P, Xing L, Hong X, et al. LncRNA PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA. Cancer Med 2020;9:7706-16. [Crossref] [PubMed]
- Xu Y, Li J, Wang P, et al. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-κB. J Cell Biochem 2019;120:19415-21. [Crossref] [PubMed]
- Gao L, Guo YN, Zeng JH, et al. The expression, significance and function of cancer susceptibility candidate 9 in lung squamous cell carcinoma: A bioinformatics and in vitro investigation. Int J Oncol 2019;54:1651-64. [Crossref] [PubMed]
- Liu L, Li Y, Zhang R, et al. MIR205HG acts as a ceRNA to expedite cell proliferation and progression in lung squamous cell carcinoma via targeting miR-299-3p/MAP3K2 axis. BMC Pulm Med 2020;20:163. [Crossref] [PubMed]
- Lu YB, Jiang Q, Yang MY, et al. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget 2017;8:88804-14. [Crossref] [PubMed]
- Wang Q, Yang L, Hu X, et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget 2017;8:3441-53. [Crossref] [PubMed]
- Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288-300. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Joshi A, Zanwar S, Noronha V, et al. EGFR mutation in squamous cell carcinoma of the lung: does it carry the same connotation as in adenocarcinomas? Onco Targets Ther 2017;10:1859-63. [Crossref] [PubMed]
- Liao RG, Watanabe H, Meyerson M, et al. Targeted therapy for squamous cell lung cancer. Lung Cancer Manag 2012;1:293-300. [Crossref] [PubMed]
- Derman BA, Mileham KF, Bonomi PD, et al. Treatment of advanced squamous cell carcinoma of the lung: a review. Transl Lung Cancer Res 2015;4:524-32. [PubMed]
- Li L, Yin JY, He FZ, et al. Long noncoding RNA SFTA1P promoted apoptosis and increased cisplatin chemosensitivity via regulating the hnRNP-U-GADD45A axis in lung squamous cell carcinoma. Oncotarget 2017;8:97476-89. [Crossref] [PubMed]
- Li G, Wang X, Luo Q, et al. Identification of key genes and long non-coding RNAs in celecoxib-treated lung squamous cell carcinoma cell line by RNA-sequencing. Mol Med Rep 2018;17:6456-64. [Crossref] [PubMed]
- Moran C. Importance of molecular features of non-small cell lung cancer for choice of treatment. Am J Pathol 2011;178:1940-8. [Crossref] [PubMed]
- Lu T, Wang Y, Chen D, et al. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther 2018;11:8045-52. [Crossref] [PubMed]
- Bolha L, Ravnik-Glavač M, Glavač D. Long Noncoding RNAs as Biomarkers in Cancer. Dis Markers 2017;2017:7243968. [Crossref] [PubMed]
- Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers 2016;2016:9085195. [Crossref] [PubMed]
- Lemos AEG, Matos ADR, Ferreira LB, et al. The long non-coding RNA PCA3: an update of its functions and clinical applications as a biomarker in prostate cancer. Oncotarget 2019;10:6589-603. [Crossref] [PubMed]
- Wu Y, Li L, Wang Q, et al. LINC00511 promotes lung squamous cell carcinoma proliferation and migration via inhibiting miR-150-5p and activating TADA1. Transl Lung Cancer Res 2020;9:1138-48. [Crossref] [PubMed]
- Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol 2015;36:7465-71. [Crossref] [PubMed]
- He W, Qiao ZX, Ma B. Long noncoding RNA FAM201A mediates the metastasis of lung squamous cell cancer via regulating ABCE1 expression. Eur Rev Med Pharmacol Sci 2019;23:10343-53. [PubMed]
- Wang C, Jing Q. Non-coding RNAs as biomarkers for acute myocardial infarction. Acta Pharmacol Sin 2018;39:1110-9. [Crossref] [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [Crossref] [PubMed]
- Hu J, Xu L, Shou T, et al. Systematic analysis identifies three-lncRNA signature as a potentially prognostic biomarker for lung squamous cell carcinoma using bioinformatics strategy. Transl Lung Cancer Res 2019;8:614-35. [Crossref] [PubMed]
- Huang H, Wang Y, Li Q, et al. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett 2019;446:81-9. [Crossref] [PubMed]
- Zhao Z, Liu W, Li J. miR-140-5p inhibits cell proliferation and invasion in colorectal carcinoma by targeting SOX4. Oncol Lett 2019;17:2215-20. [PubMed]
- Zhou Y, Wang B, Wang Y, et al. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett 2019;17:3835-41. [Crossref] [PubMed]
- Zou MX, Huang W, Wang XB, et al. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int J Clin Exp Pathol 2014;7:4877-85. [PubMed]
- Sui J, Xu SY, Han J, et al. Integrated analysis of competing endogenous RNA network revealing lncRNAs as potential prognostic biomarkers in human lung squamous cell carcinoma. Oncotarget 2017;8:65997-6018. [Crossref] [PubMed]
- Cheng Z, Bai Y, Wang P, et al. Identification of long noncoding RNAs for the detection of early stage lung squamous cell carcinoma by microarray analysis. Oncotarget 2017;8:13329-37. [Crossref] [PubMed]
- Malhotra J, Malvezzi M, Negri E, et al. Risk factors for lung cancer worldwide. Eur Respir J 2016;48:889-902. [Crossref] [PubMed]
- Park YR, Bae SH, Ji W, et al. GAB2 Amplification in Squamous Cell Lung Cancer of Non-Smokers. J Korean Med Sci 2017;32:1784-91. [Crossref] [PubMed]
- Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer 2012;131:2724-32. [Crossref] [PubMed]
- Buro-Auriemma LJ, Salit J, Hackett NR, et al. Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum Mol Genet 2013;22:4726-38. [Crossref] [PubMed]
- Wang TH, Hsia SM, Shih YH, et al. Association of Smoking, Alcohol Use, and Betel Quid Chewing with Epigenetic Aberrations in Cancers. Int J Mol Sci 2017;18:1210. [Crossref] [PubMed]
- Sundar IK, Nevid MZ, Friedman AE, et al. Cigarette smoke induces distinct histone modifications in lung cells: implications for the pathogenesis of COPD and lung cancer. J Proteome Res 2014;13:982-96. [Crossref] [PubMed]
- Li J, Li H, Zhang C, et al. Integrative analysis of genomic alteration, immune cells infiltration and prognosis of lung squamous cell carcinoma (LUSC) to identify smoking-related biomarkers. Int Immunopharmacol 2020;89:107053. [Crossref] [PubMed]
- Li S, Sun X, Miao S, et al. Differential protein-coding gene and long noncoding RNA expression in smoking-related lung squamous cell carcinoma. Thorac Cancer 2017;8:672-81. [Crossref] [PubMed]
- Desrichard A, Kuo F, Chowell D, et al. Tobacco Smoking-Associated Alterations in the Immune Microenvironment of Squamous Cell Carcinomas. J Natl Cancer Inst 2018;110:1386-92. [Crossref] [PubMed]
- Jaspers I. Cigarette smoke effects on innate immune mechanisms in the nasal mucosa. Potential effects on the microbiome. Ann Am Thorac Soc 2014;11:S38-42. [Crossref] [PubMed]
- Wang Y, Guo J. Immune cell landscape analysis reveals prognostic immune cells and its potential mechanism in squamous cell lung carcinoma. PeerJ 2020;8:e9996. [Crossref] [PubMed]
- Jiang T, Shi J, Dong Z, et al. Genomic landscape and its correlations with tumor mutational burden, PD-L1 expression, and immune cells infiltration in Chinese lung squamous cell carcinoma. J Hematol Oncol 2019;12:75. [Crossref] [PubMed]
- Rocha P, Hardy-Werbin M, Naranjo D, et al. CD103+CD8+ Lymphocytes Characterize the Immune Infiltration in a Case With Pseudoprogression in Squamous NSCLC. J Thorac Oncol 2018;13:e193-6. [Crossref] [PubMed]
- Xiang H, Ramil CP, Hai J, et al. Cancer-Associated Fibroblasts Promote Immunosuppression by Inducing ROS-Generating Monocytic MDSCs in Lung Squamous Cell Carcinoma. Cancer Immunol Res 2020;8:436-50. [Crossref] [PubMed]
- Boussiotis VA. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med 2016;375:1767-78. [Crossref] [PubMed]
- Larsen TV, Hussmann D, Nielsen AL. PD-L1 and PD-L2 expression correlated genes in non-small-cell lung cancer. Cancer Commun (Lond) 2019;39:30. [Crossref] [PubMed]
- Duan J, Cui L, Zhao X, et al. Use of Immunotherapy With Programmed Cell Death 1 vs Programmed Cell Death Ligand 1 Inhibitors in Patients With Cancer: A Systematic Review and Meta-analysis. JAMA Oncol 2020;6:375-84. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Huang Y, Kim BYS, Chan CK, et al. Improving immune-vascular crosstalk for cancer immunotherapy. Nat Rev Immunol 2018;18:195-203. [Crossref] [PubMed]
- Phillips T, Simmons P, Inzunza HD, et al. Development of an automated PD-L1 immunohistochemistry (IHC) assay for non-small cell lung cancer. Appl Immunohistochem Mol Morphol 2015;23:541-9. [Crossref] [PubMed]
- McLaughlin J, Han G, Schalper KA, et al. Quantitative Assessment of the Heterogeneity of PD-L1 Expression in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:46-54. [Crossref] [PubMed]
- Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol 2016;27:1492-504. [Crossref] [PubMed]
- Lanitis E, Dangaj D, Irving M, et al. Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann Oncol 2017;28:xii18-32. [Crossref] [PubMed]
- Hong L, Negrao MV, Dibaj SS, et al. Programmed Death-Ligand 1 Heterogeneity and Its Impact on Benefit From Immune Checkpoint Inhibitors in NSCLC. J Thorac Oncol 2020;15:1449-59. [Crossref] [PubMed]
- Heskamp S, Hobo W, Molkenboer-Kuenen JD, et al. Noninvasive Imaging of Tumor PD-L1 Expression Using Radiolabeled Anti-PD-L1 Antibodies. Cancer Res 2015;75:2928-36. [Crossref] [PubMed]
- Bensch F, van der Veen EL, Lub-de Hooge MN, et al. 89Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nat Med 2018;24:1852-8. [Crossref] [PubMed]
- Wu J, Xu C, Guan X, et al. Comprehensive analysis of tumor microenvironment and identification of an immune signature to predict the prognosis and immunotherapeutic response in lung squamous cell carcinoma. Ann Transl Med 2021;9:569. [Crossref] [PubMed]
- Barrera L, Montes-Servín E, Barrera A, et al. Cytokine profile determined by data-mining analysis set into clusters of non-small-cell lung cancer patients according to prognosis. Ann Oncol 2015;26:428-35. [Crossref] [PubMed]
- Shiels MS, Pfeiffer RM, Hildesheim A, et al. Circulating inflammation markers and prospective risk for lung cancer. J Natl Cancer Inst 2013;105:1871-80. [Crossref] [PubMed]
- Presley CJ, Tang D, Soulos PR, et al. Association of Broad-Based Genomic Sequencing With Survival Among Patients With Advanced Non-Small Cell Lung Cancer in the Community Oncology Setting. JAMA 2018;320:469-77. [Crossref] [PubMed]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938-45. [Crossref] [PubMed]
- Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. [Crossref] [PubMed]
- Hou J, Zhong Q. A novel immunogenomic prognostic signature in lung squamous carcinoma. Medicine (Baltimore) 2021;100:e24073. [Crossref] [PubMed]
- Kamphorst AO, Pillai RN, Yang S, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 2017;114:4993-8. [Crossref] [PubMed]
- Conway EM, Pikor LA, Kung SH, et al. Macrophages, Inflammation, and Lung Cancer. Am J Respir Crit Care Med 2016;193:116-30. [Crossref] [PubMed]
- Dai F, Liu L, Che G, et al. The number and microlocalization of tumor-associated immune cells are associated with patient's survival time in non-small cell lung cancer. BMC Cancer 2010;10:220. [Crossref] [PubMed]
- Tseng YH, Ho HL, Lai CR, et al. PD-L1 Expression of Tumor Cells, Macrophages, and Immune Cells in Non-Small Cell Lung Cancer Patients with Malignant Pleural Effusion. J Thorac Oncol 2018;13:447-53. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Faruki H, Mayhew GM, Serody JS, et al. Lung Adenocarcinoma and Squamous Cell Carcinoma Gene Expression Subtypes Demonstrate Significant Differences in Tumor Immune Landscape. J Thorac Oncol 2017;12:943-53. [Crossref] [PubMed]
- Li B, Cui Y, Diehn M, et al. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non-Small Cell Lung Cancer. JAMA Oncol 2017;3:1529-37. [Crossref] [PubMed]
- Song P, Guo L, Li W, et al. Clinicopathologic Correlation With Expression of PD-L1 on Both Tumor Cells and Tumor-infiltrating Immune Cells in Patients With Non-Small Cell Lung Cancer. J Immunother 2019;42:23-8. [Crossref] [PubMed]
- Song Q, Shang J, Yang Z, et al. Identification of an immune signature predicting prognosis risk of patients in lung adenocarcinoma. J Transl Med 2019;17:70. [Crossref] [PubMed]
- Uribesalgo I, Hoffmann D, Zhang Y, et al. Apelin inhibition prevents resistance and metastasis associated with anti-angiogenic therapy. EMBO Mol Med 2019;11:e9266. [Crossref] [PubMed]
- Berta J, Kenessey I, Dobos J, et al. Apelin expression in human non-small cell lung cancer: role in angiogenesis and prognosis. J Thorac Oncol 2010;5:1120-9. [Crossref] [PubMed]
- Zhou Y, Zhao M, Du Y, et al. MicroRNA-195 suppresses the progression of lung adenocarcinoma by directly targeting apelin. Thorac Cancer 2019;10:1419-30. [Crossref] [PubMed]
- Hibi M, Kaneda H, Tanizaki J, et al. FGFR gene alterations in lung squamous cell carcinoma are potential targets for the multikinase inhibitor nintedanib. Cancer Sci 2016;107:1667-76. [Crossref] [PubMed]
- Quintanal-Villalonga Á, Carranza-Carranza A, Meléndez R, et al. Prognostic Role of the FGFR4-388Arg Variant in Lung Squamous-Cell Carcinoma Patients With Lymph Node Involvement. Clin Lung Cancer 2017;18:667-674.e1. [Crossref] [PubMed]
- Quintanal-Villalonga A, Molina-Pinelo S, Yagüe P, et al. FGFR4 increases EGFR oncogenic signaling in lung adenocarcinoma, and their combined inhibition is highly effective. Lung Cancer 2019;131:112-21. [Crossref] [PubMed]
- Zeng R, Han M, Luo Y, et al. Role of Sema4C in TGF-β1-induced mitogen-activated protein kinase activation and epithelial-mesenchymal transition in renal tubular epithelial cells. Nephrol Dial Transplant 2011;26:1149-56. [Crossref] [PubMed]
- Li J, Wang Q, Wen R, et al. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med 2015;19:2793-805. [Crossref] [PubMed]
- Fan T, Lu Z, Liu Y, et al. A Novel Immune-Related Seventeen-Gene Signature for Predicting Early Stage Lung Squamous Cell Carcinoma Prognosis. Front Immunol 2021;12:665407. [Crossref] [PubMed]
- Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: External Validation of the Revision of the TNM Stage Groupings in the Eighth Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 2017;12:1109-21.
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 2016;8:a019505. [Crossref] [PubMed]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178-86. [Crossref] [PubMed]
- Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet 2016;17:630-41. [Crossref] [PubMed]
- Suzuki A, Makinoshima H, Wakaguri H, et al. Aberrant transcriptional regulations in cancers: genome, transcriptome and epigenome analysis of lung adenocarcinoma cell lines. Nucleic Acids Res 2014;42:13557-72. [Crossref] [PubMed]
- Yao L, Shen H, Laird PW, et al. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol 2015;16:105. [Crossref] [PubMed]
- Li XS, Nie KC, Zheng ZH, et al. Molecular subtypes based on DNA methylation predict prognosis in lung squamous cell carcinoma. BMC Cancer 2021;21:96. [Crossref] [PubMed]
- Zhang J, Luo L, Dong J, et al. A prognostic 11-DNA methylation signature for lung squamous cell carcinoma. J Thorac Dis 2020;12:2569-82. [Crossref] [PubMed]
- Zhang W, Cui Q, Qu W, et al. TRIM58/cg26157385 methylation is associated with eight prognostic genes in lung squamous cell carcinoma. Oncol Rep 2018;40:206-16. [Crossref] [PubMed]
- Wang HL, Li KZ, Li JL, et al. Prognostic value of AKAP13 methylation and expression in lung squamous cell carcinoma. Biomark Med 2020;14:503-12. [Crossref] [PubMed]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell 2007;128:669-81. [Crossref] [PubMed]
- Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol 2018;51:116-28. [Crossref] [PubMed]
- Sugimoto N, Maehara K, Yoshida K, et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res 2015;43:5898-911. [Crossref] [PubMed]
- Chen R, Hong Q, Jiang J, et al. AGTR1 promoter hypermethylation in lung squamous cell carcinoma but not in lung adenocarcinoma. Oncol Lett 2017;14:4989-94. [Crossref] [PubMed]
- Ni S, Ye M, Huang T. Short stature homeobox 2 methylation as a potential noninvasive biomarker in bronchial aspirates for lung cancer diagnosis. Oncotarget 2017;8:61253-63. [Crossref] [PubMed]
- Guo H, Zhou S, Tan L, et al. Clinicopathological significance of WIF1 hypermethylation in NSCLC, a meta-analysis and literature review. Oncotarget 2017;8:2550-7. [Crossref] [PubMed]
- Kim DS, Lee WK, Park JY. Promoter methylation of Wrap53α, an antisense transcript of p53, is associated with the poor prognosis of patients with non-small cell lung cancer. Oncol Lett 2018;16:5823-8. [Crossref] [PubMed]
- Zhang X, Yang X, Wang J, et al. Down-regulation of PAX6 by promoter methylation is associated with poor prognosis in non small cell lung cancer. Int J Clin Exp Pathol 2015;8:11452-7. [PubMed]
- Han P, Liu Q, Xiang J. Monitoring methylation-driven genes as prognostic biomarkers in patients with lung squamous cell cancer. Oncol Lett 2020;19:707-16. [PubMed]
- Li R, Yin YH, Jin J, et al. Integrative analysis of DNA methylation-driven genes for the prognosis of lung squamous cell carcinoma using MethylMix. Int J Med Sci 2020;17:773-86. [Crossref] [PubMed]
- Zhu Q, Wang J, Zhang Q, et al. Methylation-driven genes PMPCAP1, SOWAHC and ZNF454 as potential prognostic biomarkers in lung squamous cell carcinoma. Mol Med Rep 2020;21:1285-95. [Crossref] [PubMed]
- Zhang L, Peng D, Sood AK, et al. Shedding Light on the Dark Cancer Genomes: Long Noncoding RNAs as Novel Biomarkers and Potential Therapeutic Targets for Cancer. Mol Cancer Ther 2018;17:1816-23. [Crossref] [PubMed]
- Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med 2018;24:257-77. [Crossref] [PubMed]
- Kjems J, Howard KA. Oligonucleotide delivery to the lung: waiting to inhale. Mol Ther Nucleic Acids 2012;1:e1. [Crossref] [PubMed]
- Schoenfeld AJ, Hellmann MD. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020;37:443-55. [Crossref] [PubMed]
- Ishizuka JJ, Manguso RT, Cheruiyot CK, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019;565:43-8. [Crossref] [PubMed]
- Cortez MA, Masrorpour F, Ivan C, et al. Bone morphogenetic protein 7 promotes resistance to immunotherapy. Nat Commun 2020;11:4840. [Crossref] [PubMed]
- Kim HJ, Cantor H, Cosmopoulos K. Overcoming Immune Checkpoint Blockade Resistance via EZH2 Inhibition. Trends Immunol 2020;41:948-63. [Crossref] [PubMed]
- Lin EW, Karakasheva TA, Lee DJ, et al. Comparative transcriptomes of adenocarcinomas and squamous cell carcinomas reveal molecular similarities that span classical anatomic boundaries. PLoS Genet 2017;13:e1006938. [Crossref] [PubMed]
- Chakravarthi BV, Nepal S, Varambally S. Genomic and Epigenomic Alterations in Cancer. Am J Pathol 2016;186:1724-35. [Crossref] [PubMed]
- Huang T, Chen X, Hong Q, et al. Meta-analyses of gene methylation and smoking behavior in non-small cell lung cancer patients. Sci Rep 2015;5:8897. [Crossref] [PubMed]
- Pfeifer GP. Defining Driver DNA Methylation Changes in Human Cancer. Int J Mol Sci 2018;19:1166. [Crossref] [PubMed]
- Tan SX, Hu RC, Tan YL, et al. Promoter methylation-mediated downregulation of PRDM5 contributes to the development of lung squamous cell carcinoma. Tumour Biol 2014;35:4509-16. [Crossref] [PubMed]
- Wang W, Qi X, Wu M. Effect of DR4 promoter methylation on the TRAIL-induced apoptosis in lung squamous carcinoma cell. Oncol Rep 2015;34:2115-25. [Crossref] [PubMed]
- Doroshow DB, Sanmamed MF, Hastings K, et al. Immunotherapy in Non-Small Cell Lung Cancer: Facts and Hopes. Clin Cancer Res 2019;25:4592-602. [Crossref] [PubMed]
- Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019;322:764-74. [Crossref] [PubMed]
- Carlisle JW, Steuer CE, Owonikoko TK, et al. An update on the immune landscape in lung and head and neck cancers. CA Cancer J Clin 2020;70:505-17. [Crossref] [PubMed]


