
Patient-derived tumor spheroid cultures as a promising tool to assist personalized therapeutic decisions in breast cancer
Introduction
Breast cancer is the most common cause of cancer related deaths for women worldwide. In Europe alone, approximately 630,000 women died from breast cancer in 2018 (1). According to current European and American guidelines for treatment, breast cancers are categorized into molecular subtypes based on the expression of hormone receptors PR and ER as well as HER2 and Ki67. The subtypes include luminal A, luminal B, HER2+/non-luminal, and basal-like/triple negative, and each subtype has its own course of treatment, with endocrine therapy recommended for hormone receptor positive patients and anti-HER2 drugs for patients with high levels of HER2 expression (1,2). Yet, for the basal-like/triple negative subtype, where neither endocrine therapy nor anti-HER2 therapy is appropriate, the first-line of therapy is classic chemotherapy which includes anthracyclines and/or taxanes (2-4).
While in breast cancer there is a clear protocol for choosing a treatment plan for each patient, this treatment plan is, unfortunately, not always successful. Nearly 40% of women acquire tamoxifen resistance. Furthermore, intrinsically tamoxifen-resistant cancers often have a large number of gene alterations, making the choice of an alternative therapy anything but straightforward (5,6). In addition, there are no effective biomarkers to guide the choice of chemotherapy from a number of approved regimens. An unguided choice for first-line therapy can lead to a delay in effective treatment, and thus risk progression of the disease. Furthermore, each course of treatment is accompanied by suffering due to adverse side effects of chemotherapy (7,8). Even when actionable biomarkers do exist, they are not universally applicable and some patients who express a biomarker that is correlated with drug response do not respond to the predicted therapy. New tools to predict drug efficacy for individual patients would extend survival and prevent treatment with ineffective drugs.
One approach to developing new tools to guide individualized treatment selection utilizes chemotherapy sensitivity and resistance assays (CSRAs) which use viable tissue from a tumor to provide predictive information about response to treatment (9,10). A number of three dimensional (3D) models have been developed including tumor cells seeded in a matrix of extracellular proteins, multicellular tumor spheroids, organoids, tissue slices, bioreactors and microfluidic models (10-18). The most efficient system to quickly and inexpensively culture patient-derived tissue in a 3D model are spheroids, scaffold-free, multicellular spheres containing cancer cells. These spheroids enable cell-cell interactions and can facilitate the production of the endogenous extracellular matrix to provide local tumor microenvironment-like conditions. Additional environmental considerations, including the access to nutrients, oxygen, growth factors, metabolites and paracrine factors, are also recapitulated (11,18). Unlike organoids, there is no need for expensive reagents such as specialized cell media factors or a basement membrane matrix (such as Matrigel) that can complicate downstream analysis of the tissue. Recently, it was shown that patient-derived spheroids could be grown from ovarian tumors and that these spheroids could accurately predict response to first-line therapy with 89% accuracy (19). This clearly demonstrates the potential of this method to facilitate treatment choice, as well as to explore non-standard therapies, both of which would prevent a delay in effective treatment, eliminate unnecessary suffering, and ultimately improve prognosis.
To date, spheroids have been grown from a variety of breast cancer cell lines (20-24). Yet, successfully growing spheroids from breast cancer patient samples has proven to be a challenge. This is due to a number of factors including the small size of breast tumors, the low number of viable cells which can be extracted and the heterogeneity of breast tissue which includes a high level of non-malignant tissue including adipose tissue (25,26). We have only seen two publications from a single research group where spheroids have been successfully grown from patient tumor tissue (26,27).
Here, we present a successful method to grow in vitro spheroids from patient-derived breast tumor tissue. Our method is low cost, suited to drug screening, rapid and applicable to a large variety of breast cancer subtypes with nearly 90% success. This represents a feasible model for drug efficacy prediction for breast cancer patients that can help guide treatment decision making.
We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1577/rc).
Methods
Ethical approval and collection of patient samples
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for the research was obtained from the Institutional Helsinki Committee at the Barzilai University Medical Center in Ashkelon, Israel according to the ICH-GCP guidelines (protocol No. 0282-18-TLV). Informed written consent to have data/samples from their medical records used in research, was obtained from all participants and/or their legal guardians and all research was performed in accordance with relevant guidelines and regulations. All tissue samples were collected between August 2018 and December 2019. Criteria for subject inclusion included women over the age of 18 with a pathology assessment validating breast cancer of stage IA–IIA (based on AJCC staining version 7) who underwent surgical removal of a primary tumor (either local or mastectomy) with available data on the ER, PR, and HER2 expression status from a preliminary core needle biopsy. Samples were fully anonymized by the Institutional Biobank at Tel Aviv Sourasky Medical Center, Israel directly after surgery and were transported to our laboratory on ice in sterile tubes containing HypoThermosol® FRS medium (StemCell Technologies, Vancouver, Canada). Tissue was processed within 72 h of surgery. Sample and clinical information about the patient were acquired anonymously, with identifying information encoded at the clinical site. Medical records for retrospective study from selected patients were received 6 months after collection.
Generation of spheroids
The tissue was placed in a sterile 10 cm dish, photographed, and cut into 1–3 mm3 pieces using a sterile razor blade. Representative pieces were saved for immunohistochemistry (IHC) analysis of the original tissue. The rest was further minced. Eventually, all remaining tissue was placed in a falcon tube containing Advanced DMEM supplemented with 1% Glutamax, 1% HEPES 1M, 1% Pen/Strep (all Gibco, Waltham, MA, USA), 1× primocin (InvivoGen, San Diego, CA, USA), 0.5 mg/mL collagenase (Sigma, Burlington, MA, USA) and 0.2 mg/mL DNAse I (Sigma), and incubated on an orbital shaker at 220 rpm at 37 °C for up to 16 h. Where necessary, an additional digestion with 1× TrypLE (Gibco) was performed for 10 min at 37 °C. Then, red blood cells were eliminated using the BD Pharm LyseTM Lysing Buffer (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. Finally, the cell suspension was strained over 40 µm Corning® cell strainers (Corning, New York, NY, USA), and the single cell yield was assessed using the LUNATM (Logos Biosystems, Anyang, South Korea) cell counter. Spheroids were generated using either the 3D InSightTM (GravityTRAPTM; GravityPLUSTM) Hanging Drop System (InSphero AG, Schlieren, Switzerland) or the Corning® ULA spheroid microplates (Corning CLS4515) according to the manufacturer’s instructions. A total of 1,000 patient-derived cells were seeded in co-culture with HDFs (Sigma 106-05A) in a ratio of 1:1 or 1:3 per well in a 96-well plate format. Spheroids were grown in Mammary Epithelial Basal Medium (Lonza, Basel, Switzerland) supplemented with 2 mM L-Glutamine, 1% Penicillin-Streptomycin, 1% Fetal Bovine Serum (all Gibco), 5 µg/mL insulin (PromoCell, Heidelberg, Germany), 0.5 µg/mL Hydrocortisone, 20 ng/mL EGF, 20 ng/mL human FGF10, 1 U/mL Heparin, 50 µM L-Ascorbic acid, 50 ng/mL Cholera Toxin, 35 µg/mL BPE (all Sigma), 1× B27 supplement (Life Technologies, San Diego, CA, USA) and 20 ng/mL beta-Estradiol (Sigma). Media was changed every 2–3 days and spheroid growth was monitored by light microscopy using the Zeiss Axio Observer fluorescent microscope. The diameter “d” of spheroids was determined using the measurement feature of ImageJ (Version 1.52a). The size of each spheroid was calculated as: area = π * (d/2)2.
Histology and IHC
Spheroids and original tissue pieces were collected into 1.5 mL Eppendorf tubes, washed in 1× PBS and fixed with 4% Paraformaldehyde (Bar Naor, Ltd., Petah Tikva, Israel) for 15 min at RT. All further histology and IHC steps were performed by the staff of PathoLab (Rehovot, Israel). H&E staining was done using the Tissue Tek Prisma device under standard conditions. IHC staining for specific markers was done using the Ventana BenchMark Ultra System. The antibodies that were used and the respective dilutions can be found in the supplementary information online (Table S1).
For the panCK analysis, spheroids generated from one patient were stained as described above and the images were segmented using the Weka Segmentation plugin from ImageJ (28). Three classes were generated: background, panCK positive and spheroid. These classes were used to train a classifier and this classifier allowed us to identify the integrated density of the panCK positive region and the integrated density of the total spheroid region. Percent panCK was the panCK positive region/the total spheroid region.
Drug panel and CellTiter-Glo® 3D Cell Viability Assay
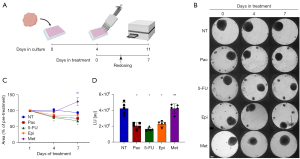
Breast cancer patient-derived spheroids were cultured in InSphero 3D InSightTM plates for 4 days. The following compounds were applied in 5 replicates per treatment at the respective final concentration: 100 nM Pac (T7191), 384 µM 5-FU (F6627), 1 µM Epi (E9406), 20 Mm Met (PHR1084, all Sigma Aldrich). Redosing was performed after 3 days, and viability of the spheroids was determined after 7 days of treatment with the CellTiter-Glo® 3D Cell Viability Assay (Promega G9682) according to the manufacturer’s instructions. Luminescence readout was read in CLARIOstar® Plus plate reader (BMG LABTECH).
Statistical analysis
The growth curve of spheroids was determined from the spheroid area. The data is presented as percentage of the size on day 1 of treatment; 5 replicates were measured. Student’s t-test and two-way ANOVA followed by Dunnett’s test were performed for statistical analysis. Graphs showing mean values with SEM were generated using GraphPad Prism {Version 9.0.0 [121]}.
For the cell viability, assay the mean of absolute luminescence from 5 replicates was determined. Student’s t-test and two-way ANOVA followed by Dunnett’s test were performed for statistical analysis. Graphs showing individual values, and mean values with SEM were generated using GraphPad Prism {Version 9.0.0 [121]}.
Availability of data and materials
All clinical data from all patient-derived samples are included in this published article and can be found in the supplementary information online (Table S2).
All generated or analyzed data sets in this study are available from the corresponding author on reasonable request.
Results
Generation of spheroids from surgical samples of human breast cancer tissue
In order to establish a working protocol to grow in vitro spheroids from human patient material, tissue samples were received post-surgery from previously untreated human female breast cancer patients (Figure 1). After dissociation of the tissue into a single cell suspension, spheroids were generated using a scaffold-free approach in the 96-well plate format from InSphero 3D InSightTM (GravityTRAPTM and GravityPLUSTM) Hanging Drop System or the Corning® ultra-low attachment (ULA) spheroid microplates. We observed that the reconstitution of the tumor microenvironment with stromal cells using normal human fibroblasts was fundamental to ensure the generation of multicellular aggregates into stable tumor spheroids. This approach was based on previous work with breast cancer cell lines which also used normal human dermal fibroblasts (HDFs) (20), and is also in line with the fact that fibroblasts play an important role in signaling cascades in tumor cell proliferation (29). The best results were achieved with a tumor cell-to-fibroblast ratio of 1:3 (Figure 2A), but a 1:1 ratio also produced smooth, round and tightly packed spheroids (Figure 2B,2C). The fibroblasts are recognized by the epithelial cells and serve as a scaffold to facilitate the self-assembly of the tumor cells.
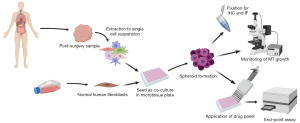
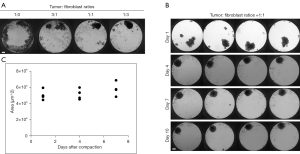
On average, complete cell agglomeration into compact spheroids was seen after two to four days under optimal culture conditions (Figure 2B). The size variation between individual replicates of one patient sample is very low (Figure 2B,2C). Once formed, spheroids could be fixed and stained for IHC or immunofluorescence (IF), monitored for growth by light microscopy and/or cell viability assays, or treated with a panel of chemotherapy drugs (Figure 1).
Spheroids can be successfully grown across breast cancer tissues with distinct molecular signatures
We received surgical samples from a cohort of 31 breast cancer patients with varying genetic backgrounds and tumor grades. It is important to note that we were able to successfully grow spheroids from 27 out of the 31 samples, with an overall success rate of 87%. Our established criterion for success was the ability to grow at least 10 spheroids from the tumor tissue which would give us enough spheroids for control spheroids and drug testing of two different treatments. In 21 samples we were able to grow 20 spheroids and above which enabled us to test five different drugs or combinations. We had access to pathological and clinical data for most of the original tissue material. Table 1 shows a summary of the samples used in the study with the available information regarding tumor stage and genetic background, as well as the success rate of establishing spheroids from the given tissues. We successfully grew spheroids from all genetic backgrounds and tumor grades received which included all possible genetic backgrounds for breast cancer with the exception of the HER2+/non-luminal type which we did not receive from the biobank. Spheroid growth success rate was independent of these tumor features based on the intrinsic data set. It is important to note that all samples from the triple-negative subtype were successfully grown using this procedure. This is encouraging since there is a lack of targeted drugs for this particular subtype (1,30-32). Thus, our established working protocol can generate in vitro spheroids from breast cancer tissues of different stages and genetic backgrounds.
Table 1
| Features of tumor samples | Total number of samples | Number of successful samples |
|---|---|---|
| Tumor stage | ||
| I–II | 4 | 4 (100%) |
| II | 9 | 7 (78%) |
| II–III | 4 | 4 (100%) |
| III | 5 | 4 (80%) |
| N/A | 9 | 8 (88%) |
| Total | 31 | 27 (87%) |
| Genetic background | ||
| Triple negative | 4 | 4 (100%) |
| Luminal A | 11 | 10 (91%) |
| Luminal B1 | 10 | 8 (80%) |
| Luminal B2 | 4 | 3 (75%) |
| N/A | 2 | 2 (100%) |
| Total | 31 | 27 (87%) |
| Ki67 | ||
| 0–10% | 17 | 15 (88%) |
| 11–20% | 6 | 6 (100%) |
| 21–40% | 3 | 3 (100%) |
| N/A | 5 | 3 (60%) |
| Total | 31 | 27 (87%) |
The spheroids contain viable epithelial tumor tissue which maintains characteristics of the original tumor
The promising results achieved in generating in vitro spheroids from breast tumors of human origin prompted us to determine the cellular composition of these spheroids. For this, spheroids from one patient cultured in the InSphero 3D InSightTM or Corning® ULA microplates, were fixed with paraformaldehyde and characterized by IHC. We found that spheroids generated from breast tissue contained both epithelial cells (as shown by panCK staining) and stromal cells (indicated by vimentin staining) (Figure 3A,3B). The epithelial cells in the spheroid are of tumor tissue origin, demonstrating that our technique supports the growth of original tumor tissue in the spheroid model. As expected, spheroids that were generated from HDFs alone only stained positive for vimentin and not panCK (information can be found in supplementary information online, Figure S1). It is important to note that we did not observe vimentin-positive tumor cells. The tumor spheroids had clear Ki-67 staining indicating that the tumor-derived cells were actively proliferating in the spheroids (Figure 3C). In contrast, the HDF-only spheroids did not have Ki-67 staining (Figure 3C), showing that there is no concern that the fibroblasts proliferate and may become the bulk of the spheroid. Rather they mainly serve a structural and maintenance role. We show data from three representative spheroids from the same patient.
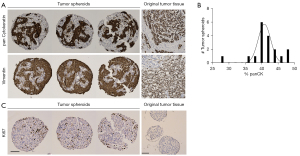
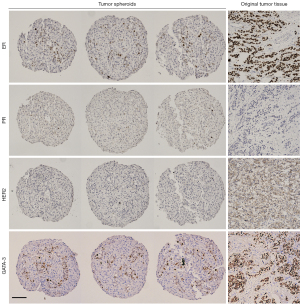
Once we showed that viable tumor cells integrate into our spheroids, we investigated whether they maintain the molecular characteristics of the original tumor. For this investigation we used the markers most commonly used in the clinic to classify breast cancer and determine treatment, ER, PR and HER2 (1,2). Figure 4 shows a representative example of the same original tumor sample used in Figure 3, identified by clinical data as ER+, PR− and HER2− (equivocal IHC, designated negative by FISH) and classified as luminal B1 (1). IHC staining of the spheroids from the same patient sample for these markers revealed that, like the original tissue, the spheroids were PR negative with low-no HER2 staining (Figure 4). There was a significant number of ER+ cells in the original tumor. In the spheroids there was a small population of ER+ cells, this distribution is expected in light of the fact that the epithelial cells represent 25–50% of the spheroid content. After establishing that the spheroids reflected the staining pattern of the original tumor tissue, we examined another breast cancer marker, GATA-3, commonly expressed in over 90% of luminal B breast cancers (33). GATA-3 was expressed in discrete cells in the original tumor and all three replicates of the tumor spheroids (Figure 4). Furthermore, we stained the HDF spheroids with the same panel of breast cancer markers and showed that they did not stain for any of the breast cancer markers (more information in the supplementary information online, Figure S1). This shows that expression of markers classically used to characterize breast cancer are maintained in the spheroid model and that this model recapitulates the original tissue in this respect.
Spheroids respond to standard chemotherapy drugs
After successfully growing breast cancer spheroids from a variety of breast cancer patients and confirming that the spheroids reflect the molecular characteristics of the original tissue, we investigated whether this model could serve as a platform to test drug sensitivity. Therefore, to test the feasibility of the 3D spheroid model as a drug screening model, breast cancer patient-derived spheroids were treated with FDA-approved drugs. The list included paclitaxel (Pac), epirubicin (Epi), 5-fluorouracil (5-FU) and metformin (Met).
Spheroids were generated using the InSphero 3D InSightTM system. Following 4 days of incubation under optimal growth conditions, spheroids were treated with Pac, Epi, 5-FU, and Met for a period of up to one week (Figure 5A). The concentration of each drug was chosen from previous studies on two dimensional (2D) models and increased in accordance with the 3D setup (34-36). Spheroid size was monitored by light microscopy, and the area was determined using ImageJ (Figure 5B). The size of the spheroids decreased significantly upon treatment with 5-FU (Figure 5C). Pac and Epi treatments also led to a visible reduction in spheroid size (Figure 5B). Following treatment, ATP levels were measured using a luminescence-based ATP assay (Figure 5D). As expected, ATP levels significantly decreased under treatment with the first- and second-line chemotherapy drugs for breast cancer, Pac, 5-FU and Epi, while Met showed no reduction in ATP levels (Figure 5D) or spheroid size (Figure 5B,5C). Thus, the patient-derived spheroids respond, with expected variability, to clinically relevant chemotherapy drugs.
Discussion
Improved method to generate spheroids from human breast cancer tissue
In this study we present an improved method for generating spheroids from cancerous human breast tissue. While spheroids have been derived from patient tissue in a number of histotypes, their generation from breast tumor tissue has proved challenging (25,26,37,38). We are aware of only two publications where breast cancer spheroids have been successfully grown from patient samples (26,39). Here, we used material derived from a cohort of breast cancer patients to establish an effective protocol for the generation of spheroids utilizing a scaffold-free approach and tested their applicability in a variety of end-point assays. We employed a generic growth media that was supplemented with necessary growth factors and additives, such as insulin, heparin and hydrocortisone, as well as beta-estradiol. However, we refrained from adding stem cell niche factors, in order to avoid selective growth of only one particular subpopulation (40). As described, stromal cells are indispensable to the 3D architecture of the tissue, contributing to the physical cell organization and to the biochemical signaling within the tumor microenvironment, and thus support key properties of solid tumors. Fostering cell-cell interactions between normal fibroblasts and cancer epithelial cells within the spheroids is therefore critically important for mimicking the tumor microenvironment in vitro (41-45). On the other hand, excessive proliferation of the fibroblasts could threaten the overall composition of the spheroid. For this reason, we carefully selected the supplements of the growth media. For example, FGF10 has been reported to selectively promote epithelial cancer cell growth, while cholera toxin reduces the expansion of fibroblasts (15,46,47). In addition, in most cases, the spheroids were cultured for a maximum of 2 weeks in medium with a low percentage of serum, thereby also limiting the expansion of the fibroblast population. Ki-67 staining of HDF and tumor spheroids demonstrated that the fibroblasts were not an actively replicating population in the spheroids (Figure 3C).
Our success rate in growing spheroids from primary tissue was at 87%, which is significantly higher than previously reported 73% and 59% for work on patient-derived spheroids in breast cancer (26,27). Spheroids which failed to grow (4 out of 31) were mostly due to contamination with bacteria or fungi (in 3 cases), or low quality of the original tissue piece which resulted in a low yield of viable epithelial cells after single cell extraction (in 1 case). This high success rate means that this system could be applicable for the vast majority of breast cancer patients.
Validation of spheroid composition
In light of the heterogeneity of breast tumor tissue it was essential to ensure that the spheroids contained epithelial tissue. Therefore, we performed IHC analysis to identify and discriminate between the epithelial and stromal cells. This type of analysis was not performed in the two studies of spheroids from patient breast cancer tissue mentioned above (26,27). It has previously been shown that in spheroids generated from co-cultures of cancer cell lines and fibroblast lines the epithelial cells are primarily localized to the periphery of the spheroid, whereas the fibroblast cells make up the core (41,48). Surprisingly, and in contrast to what we and others have seen in spheroids from co-cultured cell lines, we did not detect a distinct localization of epithelial and stromal cells in the spheroids generated from patient-derived material (see Figure 3 as a representative image of our consistent observation of this phenomenon). We also performed Ki-67 staining which showed that a portion of the epithelial cells were replicating while the fibroblast cells were not. In conclusion, our patient-derived breast cancer spheroids were composed of a mixture of fibroblasts and epithelial tumor tissue with no clear compartmentalization of the two populations.
Furthermore, we showed that the composition of the spheroids recapitulates the molecular features of the original tissue with regards to the classic breast cancer markers ER, PR and HER2 (Figure 4). Since these markers are standard in the clinic to characterize the tumor, it is important to note that their expression was maintained in the spheroid model (1,2). We further characterized the spheroids by looking at an additional common breast cancer marker, GATA-3, and saw that this marker is expressed both in the original tissue and in the tumor spheroids. This type of investigation was not performed in the previous studies on breast cancer spheroids. These data provide an important piece in the validation of our method.
Responsiveness of spheroids to drug panel
To provide a preliminary proof-of-concept that spheroids could, indeed, be utilized as in vitro models for the prediction of drug response in personalized medicine, we applied a small panel of commonly used chemotherapy and adjuvant drugs, including Pac, Epi, 5-FU, and Met onto spheroids generated from patient-derived breast cancer tissue. While the tumor sample we used was of the luminal A subtype which is most commonly treated by endocrine therapy, single agent chemotherapy is often used as a second-line therapy in these cases or if there is high tumor burden (2). Given significant levels of tamoxifen resistance in ER+ cancer (up to 40%) and recurrence of up to 22% in 5 years it is important to arm doctors with tools to determine treatment in the case of resistance or recurrence (5,6,49). In future experiments we plan to test the first-line recommendations for the tumor type on the spheroids. Our system could also be used as a low risk assay to determine individual response to more experimental drugs. For example, Met has recently been reported to provide a promising adjuvant treatment for prostate and colorectal cancer and is currently being investigated as adjuvant therapy for breast cancer in an active phase III clinical trial (50). With our drug panel we showed the potential of our system to reveal drug response for classic chemotherapy and investigational drugs.
As a readout for cell viability of the spheroids upon drug treatment, we chose a simple size assessment from bright-field images, as well as a luminescence-based ATP assay. We show that the reduction in spheroid size correlates with a decrease in intracellular ATP levels. All tested first-line chemotherapy drugs for breast cancer affected the viability of the spheroids, as expected. Similar drug panel assays in other contexts consist of dose-response data of compounds to estimate an appropriate dose for each drug regimen. Due to the relatively low yield of viable single cells extracted from breast cancer tissues, it is not possible at this time to generate enough spheroids to perform such analysis. However, in general, there are many challenges in the translation of doses determined in vitro to clinical dosing. The primary goal of a personalized medicine assay is foremost to assess the drugs and drug combinations to which an individual tumor is sensitive, rather than to provide exact dosing information.
Here, our main focus was to investigate spheroids for their variability and sensitivity to single agent application of specific drugs that were selected based on their clinical relevance for the tumor type. This valid biological platform can significantly contribute to the choice of first-line treatment where single or combinatorial drugs are used for a better therapeutic response in individual patients. Shuford et al. (19) have recently reported that spheroids generated from ovarian tissues accurately predicted the response and non-response of ovarian cancer patients to specific treatments.
Summary: benefits and limitations of the spheroid model
3D models provide enormous benefit beyond conventional 2D cell culture models with regards to establishing an efficient in vitro pre-clinical platform for drug screening that accurately represents the original tumor (10,19,44). Several 3D models have been developed including organoids, tissue slices, hydrogels, bioreactors, microfluidic models and scaffold-free spheroids (10-14,16-18,37). In our research, we investigated the utility of spheroids as a model for drug testing on patient-derived breast cancer tissue. One of the primary limitations that is true of all 3D culture methods, is the unpredictable quality of human biopsies and surgical resections as source of starting material. The tissue heterogeneity and size differ between various tissue types. Ultimately, breast tissue contains a high percentage of fat cells and a relatively low volume of epithelial cells, whereas other tissue types, such as colon or prostate yield a higher number of viable cancer cells (25). Despite the challenges presented with the starting tissue in breast cancer we were able to generate spheroids with the relatively high success rate of nearly 90%. Before this model can be firmly established as a viable system for prediction of drug efficacy a more thorough investigation of the subpopulations that exist in the original tumor and the spheroids must be performed.
Notably, the biggest advantage of our method, which utilizes the Corning® ULA microplates and the InSphero 3D InSightTM system is the straightforward seeding procedure and the ease of plate handling. In addition, within only a few days, spheroids are already visible and functional for drug treatment and respective end-point assays. Therefore, they provide a promising tool for pre-clinical prediction of drug response in personalized cancer therapy.
Overall, our success rate in generating spheroids from nearly 90% of the breast cancer tissue samples obtained, as well as the rapid time frame, use of minimal equipment and specialized reagents, and flexibility, support the potential of this method for clinical application. We were able to perform fifteen drug panels with at least one standard of care drug and we were consistently able to show a patient specific response. Our system, therefore, has the potential to be utilized as a tool for personalized prediction of the responsiveness of individual cancerous tissue to selected chemotherapeutic options. Further optimization and validation of our protocol will be performed in future pro- and retrospective studies.
Acknowledgments
We thank Olga Shovman, Nili Turian, Karin Abaev, Hadas Fulman-Levy, and Tzachi Morad for technical assistance, and Drs. Sharona Even Ram and Gabriela Rozic for comments on the manuscript (all from the Institute for Personalized and Translational Medicine). We are grateful to Drs. Francesca Chiovaro, Wolfgang Moritz (both InSphero AG) and Dr. Sumeer Dhar for helpful discussions and comments on the study and the manuscript. We also thank Dr. Ayelet Izhaki and coworkers from the Institutional Biobank at Tel Aviv Sourasky Medical Center for help in collecting surgical specimens.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1577/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1577/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1577/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Helsinki Committee at the Barzilai University Medical Center in Ashkelon, Israel according to the ICH-GCP guidelines (protocol No. 0282-18-TLV). Informed written consent to have data/samples from their medical records used in research, was obtained from all participants and/or their legal guardians and all research was performed in accordance with relevant guidelines and regulations.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cardoso F, Kyriakides S, Ohno S, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2019;30:1194-220. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017;389:2430-42. [Crossref] [PubMed]
- Yuan C, Liu Z, Yu Q, et al. Expression of PD-1/PD-L1 in primary breast tumours and metastatic axillary lymph nodes and its correlation with clinicopathological parameters. Sci Rep 2019;9:14356. [Crossref] [PubMed]
- Hultsch S, Kankainen M, Paavolainen L, et al. Association of tamoxifen resistance and lipid reprogramming in breast cancer. BMC Cancer 2018;18:850. [Crossref] [PubMed]
- Poudel P, Nyamundanda G, Patil Y, et al. Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses. NPJ Breast Cancer 2019;5:21. [Crossref] [PubMed]
- Fotheringham S, Mozolowski GA, Murray EMA, et al. Challenges and solutions in patient treatment strategies for stage II colon cancer. Gastroenterol Rep (Oxf) 2019;7:151-61. [Crossref] [PubMed]
- Jensen BV. Cardiotoxic consequences of anthracycline-containing therapy in patients with breast cancer. Semin Oncol 2006;33:S15-21. [Crossref] [PubMed]
- Burstein HJ, Mangu PB, Somerfield MR, et al. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J Clin Oncol 2011;29:3328-30. [Crossref] [PubMed]
- Morgan MM, Johnson BP, Livingston MK, et al. Personalized in vitro cancer models to predict therapeutic response: Challenges and a framework for improvement. Pharmacol Ther 2016;165:79-92. [Crossref] [PubMed]
- Brancato V, Oliveira JM, Correlo VM, et al. Could 3D models of cancer enhance drug screening? Biomaterials 2020;232:119744. [Crossref] [PubMed]
- Grinshpun A, Gavert N, Granit RZ, et al. Ev vivo organ culture as potential prioritization tool for breast cancer targeted therapy. Cancer Biol Ther 2018;19:645-8. [Crossref] [PubMed]
- Jo Y, Choi N, Kim K, et al. Chemoresistance of Cancer Cells: Requirements of Tumor Microenvironment-mimicking In Vitro Models in Anti-Cancer Drug Development. Theranostics 2018;8:5259-75. [Crossref] [PubMed]
- Majumder B, Baraneedharan U, Thiyagarajan S, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun 2015;6:6169. [Crossref] [PubMed]
- Mulholland T, McAllister M, Patek S, et al. Drug screening of biopsy-derived spheroids using a self-generated microfluidic concentration gradient. Sci Rep 2018;8:14672. [Crossref] [PubMed]
- Orditura M, Della Corte CM, Diana A, et al. Three dimensional primary cultures for selecting human breast cancers that are sensitive to the anti-tumor activity of ipatasertib or taselisib in combination with anti-microtubule cytotoxic drugs. Breast 2018;41:165-71. [Crossref] [PubMed]
- Tanigawa N, Yamaue H, Ohyama S, et al. Exploratory phase II trial in a multicenter setting to evaluate the clinical value of a chemosensitivity test in patients with gastric cancer (JACCRO-GC 04, Kubota memorial trial). Gastric Cancer 2016;19:350-60. [Crossref] [PubMed]
- Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia 2015;17:1-15. [Crossref] [PubMed]
- Shuford S, Wilhelm C, Rayner M, et al. Prospective Validation of an Ex Vivo, Patient-Derived 3D Spheroid Model for Response Predictions in Newly Diagnosed Ovarian Cancer. Sci Rep 2019;9:11153. [Crossref] [PubMed]
- Anastasov N, Höfig I, Radulović V, et al. A 3D-microtissue-based phenotypic screening of radiation resistant tumor cells with synchronized chemotherapeutic treatment. BMC Cancer 2015;15:466. [Crossref] [PubMed]
- Falkenberg N, Anastasov N, Höfig I, et al. Additive impact of HER2-/PTK6-RNAi on interactions with HER3 or IGF-1R leads to reduced breast cancer progression in vivo. Mol Oncol 2015;9:282-94. [Crossref] [PubMed]
- Falkenberg N, Höfig I, Rosemann M, et al. Three-dimensional microtissues essentially contribute to preclinical validations of therapeutic targets in breast cancer. Cancer Med 2016;5:703-10. [Crossref] [PubMed]
- Ho WY, Yeap SK, Ho CL, et al. Development of multicellular tumor spheroid (MCTS) culture from breast cancer cell and a high throughput screening method using the MTT assay. PLoS One 2012;7:e44640. [Crossref] [PubMed]
- Rahimi M, Sharifi-Zarchi A, Zarghami N, et al. Down-Regulation of miR-200c and Up-Regulation of miR-30c Target both Stemness and Metastasis Genes in Breast Cancer. Cell J 2020;21:467-78. [PubMed]
- Goldhammer N, Kim J, Timmermans-Wielenga V, et al. Characterization of organoid cultured human breast cancer. Breast Cancer Res 2019;21:141. [Crossref] [PubMed]
- Halfter K, Ditsch N, Kolberg HC, et al. Prospective cohort study using the breast cancer spheroid model as a predictor for response to neoadjuvant therapy--the SpheroNEO study. BMC Cancer 2015;15:519. [Crossref] [PubMed]
- Halfter K, Hoffmann O, Ditsch N, et al. Testing chemotherapy efficacy in HER2 negative breast cancer using patient-derived spheroids. J Transl Med 2016;14:112. [Crossref] [PubMed]
- Arganda-Carreras I, Kaynig V, Rueden C, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 2017;33:2424-6. [Crossref] [PubMed]
- Pal A, Ashworth JC, Collier P, et al. A 3D Heterotypic Breast Cancer Model Demonstrates a Role for Mesenchymal Stem Cells in Driving a Proliferative and Invasive Phenotype. Cancers (Basel) 2020;12:2290. [Crossref] [PubMed]
- Heimes AS, Schmidt M. Atezolizumab for the treatment of triple-negative breast cancer. Expert Opin Investig Drugs 2019;28:1-5. [Crossref] [PubMed]
- Jamdade VS, Sethi N, Mundhe NA, et al. Therapeutic targets of triple-negative breast cancer: a review. Br J Pharmacol 2015;172:4228-37. [Crossref] [PubMed]
- Telli ML, Gradishar WJ, Ward JH. NCCN Guidelines Updates: Breast Cancer. J Natl Compr Canc Netw 2019;17:552-5. [PubMed]
- Ni YB, Tsang JYS, Shao MM, et al. GATA-3 is superior to GCDFP-15 and mammaglobin to identify primary and metastatic breast cancer. Breast Cancer Res Treat 2018;169:25-32. [Crossref] [PubMed]
- Garbar C, Mascaux C, Giustiniani J, et al. Chemotherapy treatment induces an increase of autophagy in the luminal breast cancer cell MCF7, but not in the triple-negative MDA-MB231. Sci Rep 2017;7:7201. [Crossref] [PubMed]
- Muguruma M, Teraoka S, Miyahara K, et al. Differences in drug sensitivity between two-dimensional and three-dimensional culture systems in triple-negative breast cancer cell lines. Biochem Biophys Res Commun 2020;533:268-74. [Crossref] [PubMed]
- Zasadil LM, Andersen KA, Yeum D, et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med 2014;6:229ra43. [Crossref] [PubMed]
- Bolck HA, Corrò C, Kahraman A, et al. Tracing Clonal Dynamics Reveals that Two- and Three-dimensional Patient-derived Cell Models Capture Tumor Heterogeneity of Clear Cell Renal Cell Carcinoma. Eur Urol Focus 2021;7:152-62. [Crossref] [PubMed]
- Miyoshi H, Maekawa H, Kakizaki F, et al. An improved method for culturing patient-derived colorectal cancer spheroids. Oncotarget 2018;9:21950-64. [Crossref] [PubMed]
- Halfter K, Mayer B. Bringing 3D tumor models to the clinic - predictive value for personalized medicine. Biotechnol J 2017;12: [Crossref] [PubMed]
- Yin X, Mead BE, Safaee H, et al. Engineering Stem Cell Organoids. Cell Stem Cell 2016;18:25-38. [Crossref] [PubMed]
- Herter S, Morra L, Schlenker R, et al. A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol Immunother 2017;66:129-40. [Crossref] [PubMed]
- Kijima T, Nakagawa H, Shimonosono M, et al. Three-Dimensional Organoids Reveal Therapy Resistance of Esophageal and Oropharyngeal Squamous Cell Carcinoma Cells. Cell Mol Gastroenterol Hepatol 2019;7:73-91. [Crossref] [PubMed]
- Thoma CR, Zimmermann M, Agarkova I, et al. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv Drug Deliv Rev 2014;69-70:29-41. [Crossref] [PubMed]
- Weydert Z, Lal-Nag M, Mathews-Greiner L, et al. A 3D Heterotypic Multicellular Tumor Spheroid Assay Platform to Discriminate Drug Effects on Stroma versus Cancer Cells. SLAS Discov 2020;25:265-76. [Crossref] [PubMed]
- Whelan KA, Muir AB, Nakagawa H. Esophageal 3D Culture Systems as Modeling Tools in Esophageal Epithelial Pathobiology and Personalized Medicine. Cell Mol Gastroenterol Hepatol 2018;5:461-78. [Crossref] [PubMed]
- Hollenberg MD, Cuatrecasas P. Epidermal growth factor: receptors in human fibroblasts and modulation of action by cholera toxin. Proc Natl Acad Sci U S A 1973;70:2964-8. [Crossref] [PubMed]
- Memarzadeh S, Xin L, Mulholland DJ, et al. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell 2007;12:572-85. [Crossref] [PubMed]
- Amann A, Zwierzina M, Gamerith G, et al. Development of an innovative 3D cell culture system to study tumour--stroma interactions in non-small cell lung cancer cells. PLoS One 2014;9:e92511. [Crossref] [PubMed]
- Pan H, Gray R, Braybrooke J, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 2017;377:1836-46. [Crossref] [PubMed]
- Coyle C, Cafferty FH, Vale C, et al. Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann Oncol 2016;27:2184-95. [Crossref] [PubMed]


