
Circular RNA circTADA2A promotes the proliferation, invasion, and migration of non-small cell lung cancer cells via the miR-450b-3p/HMGN5 signaling pathway
Introduction
Non-small cell lung cancer (NSCLC), as the major type of lung cancer, accounts for at least 85% of all primary lung cancer cases (1). Owing to its rapid local invasions and/or distant metastases, NSCLC is becoming the prime cause of cancer-associated deaths worldwide (2), with poor total 5-year survival (3). Therefore, it is necessary to investigate the molecular mechanism of proliferation, invasion, and metastasis in NSCLC.
Circular RNAs (circRNAs), newly discovered noncoding RNAs (ncRNAs), have unique closed loop structures that ensure the stability of these molecules and resistance to degradation compared with their linear RNAs (4). CircRNAs have been involved in promoting the proliferation, invasion, and migration of various cancer cells (5). Increasing evidence has confirmed that circRNA transcriptional adaptor 2A (circTADA2A) can accelerate the progression of multiple cancers, including osteosarcoma, colorectal cancer, and breast cancer (6-8). Although Zhao et al. reported that propofol exhibited an anti-lung cancer effect by decreasing circTADA2A levels (9), the detailed mechanism by which circTADA2A aggravates NSCLC remains unclear.
High mobility group nucleosome binding domain 5 (HMGN5), a typical member of the HMGN protein family, is widely expressed in various human tissues (10). Recently, it has been demonstrated that the overexpression of HMGN5 promotes proliferation and invasion in some kinds of cancers, including prostate cancer, breast cancer, and bladder cancer (11-13). However, little is known about the function of HMGN5 in human NSCLC.
Previous studies have reported circTADA2A exert an inhibitory role in tumorigenesis including the colorectal cancer (7). Surprising, this study firstly found circTADA2A exert a promoting role in NSCLC metastasis and via up-regulating HMGN5 protein expression. The above speculative mechanisms are the innovation in NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2836/rc).
Methods
Clinical samples
A total of 60 paired clinical samples of NSCLC tissues and corresponding normal adjacent tissues were obtained from Zhuji People’s Hospital (Zhejiang, China). This study was approved by the Ethics Committee of Zhuji People’s Hospital (No. 2021-0915). All patients signed written informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
Normal human bronchial epithelial (16HBE) cells and human NSCLC cell lines (A549 and H1299) were purchased from ATCC (American Type Culture Collection). These cell lines were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) with 10% FBS (HyClone, Logan, UT, USA) in a 37 °C incubator containing 5% CO2.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted using TRIzol purchased from Invitrogen (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The primers for circTADA2A, miR-450b-3p, and HMGN5 were obtained from Invitrogen Bioengineering Corporation (Shanghai, China). qRT-PCR was performed as previously described (14). The relative RNA expression levels were analyzed using the 2−ΔΔCt method. Each sample was assayed in triplicate.
Transwell migration and invasion assay
Transwell migration was used to test cell invasion. Briefly, each Transwell insert was precoated with Matrigel, and then 3×105 cells in 200 µL of DMEM medium were seeded onto the upper chamber. Furthermore, 500 µL of DMEM with 10% serum was cultivated in the lower chamber. Following incubation for 24 h at 37 °C, the invaded A549 and H1299 cells were fixed and stained with 0.5% crystal violet (Beyotime Biotechnology, Beijing, China). Subsequently, photomicrographs were taken under an inverted light microscope (Leica, Germany).
Construction of cell lines stably transfected with circTADA2A and HMGN5
Well-designed small interfering RNA (siRNA) oligos of circTADA2A and HMGN5 as well as negative control siRNA as a control were generated from Shanghai Gene Tech (Shanghai, China) and then separately transfected into the cells with RNAiMAX (Invitrogen, USA) in Opti-MEM (Invitrogen, USA) for 48 h according to the manufacturer’s protocol. Transfection efficiency was assessed through a green fluorescent protein assay, and the stable cell lines were screened out by using puromycin.
Cell viability assay
The cell counting kit-8 (CCK-8) assay was used to test cell viability. Briefly, 1.0×104 transfected A549 and H1299 cells per well were seeded into 96-well plates and then cultured for 24, 48, or 72 h. Afterward, these cells were incubated with 10 µL of CCK-8 solution for 2 h. A microplate reader was used to test the absorbance of the plate at 450 nm.
Wound healing assay
The wound healing assay was used to measure cell migration. Briefly, A549 and H1299 cells were plated on 6-well plates to 90% confluence and scratched with a 200 µL pipette tip. At 0 h and 24 h after scratching, these cultured cells were washed twice with sterile PBS, and images were captured by an inverted light microscope (Leica, Germany). Next, wound closure (%) was assessed by Image-Pro Plus 5.1 software.
Western blotting
Fresh cell protein samples from each group of A549 and H1299 cells were lysed and isolated using RIPA lysis buffer (Beyotime Biotechnology, Beijing, China). The protein concentrations were quantified by the BCA assay (Beyotime Biotechnology, Beijing, China). Then, the same 30 µg per lane was separated by 10% SDS–PAGE and transferred onto PVDF membranes. Afterward, the membranes were blocked with 5% nonfat milk in TBST for 1 h at room temperature and incubated with anti-HMGN5 antibody (1:1,000, Abcam, USA) overnight at 4 °C. After being washed three times, the membranes were incubated with HRP-conjugated secondary anti-rabbit IgG (1:2,000, Beyotime Biotechnology, Beijing, China) for 1 h at room temperature and photographed with the Bio–Rad imaging system.
Dual-luciferase reporter assay
The regulatory interaction between circTADA2A and miR-450b-3p was investigated by dual-luciferase reporter assay. Plasmids based on the dual-luciferase reporter vector of circTADA2A-WT (wild-type), circTADA2A-MT (mutant-type), HMGN5-WT, and HMGN5-MT were constructed and confirmed with DNA sequencing (Shanghai GenePharma Co., Ltd., Shanghai, China). Then, these plasmids were cotransfected into A549 and H1299 cells with miR-450b-3p mimics or miRNA negative control (miR-NC) using Lipofectamine 2000 reagent (Invitrogen, Thermo Fisher Scientific, USA) according to the manufacturer’s protocol, and these cells were further cultured for 48 h. Next, a luciferase reporter assay was conducted as previously indicated (15). Experimental results were analyzed in triplicate and normalized to Renilla luciferase activity.
Statistical analyses
All experimental data are presented as the mean ± standard deviation (SD). The overall survival of NSCLC patients was determined with the Kaplan–Meier method. The difference between the two groups was examined via two-tailed Student’s t-test. P<0.05 was considered as statistically significant in this study.
Results
The expression of circTADA2A is upregulated and related to the poor prognosis of NSCLC patients
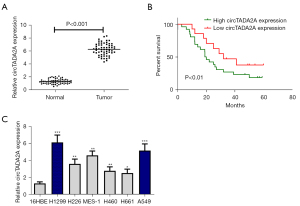
To estimate the expression of circTADA2A in human NSCLC, qRT–PCR assays were carried out in 60 paired clinical samples of NSCLC tissues and corresponding normal adjacent tissues. Compared with normal tissues, circTADA2A expression in NSCLC tissues was obviously elevated (Figure 1A). In addition, the overall survival of patients with higher expression of circTADA2A was reduced (Figure 1B). The expression of circTADA2A was upregulated in human lung cancer cell lines (H1299 and A549) relative to the normal 16HBE cell line (Figure 1C). These above findings indicate that higher expression of circTADA2A is potentially associated with poor prognosis of NSCLC patients.
CircTADA2A promotes the proliferation, invasion, and migration of NSCLC cells
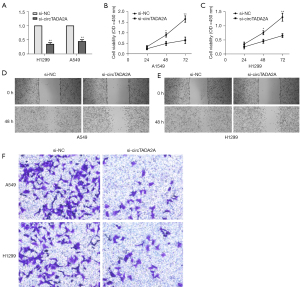
To investigate the biological effects of circTADA2A in NSCLC, a CCK-8 assay was applied to test NSCLC cell proliferation, a wound healing assay was used to assess NSCLC cell migration, and a Transwell assay was performed to evaluate NSCLC cell invasion. Specific circTADA2A siRNA successfully knocked down the expression of circTADA2A in A549 and H1299 cells (Figure 2A). In addition, circTADA2A siRNA not only successfully inhibited cell proliferation (Figure 2B,2C) but also evidently suppressed cell migration (Figure 2D,2E) and invasion (Figure 2F). Notably, circTADA2A siRNA prevented the carcinogenic behaviors of both A549 and H1299 cells. Overall, these findings suggest that circTADA2A promotes the proliferation, invasion, and metastasis of NSCLC cells in vitro.
CircTADA2A promotes NSCLC cell proliferation, invasion, and migration by upregulating HMGN5 expression
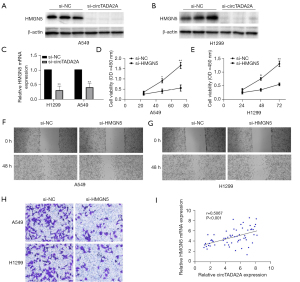
HMGN5 has been involved in promoting the proliferation, invasion, and migration of various cancer cells (11,12,16). To investigate the effect of circTADA2A on the expression of HMGN5, A549 and H1299 cells were transfected with si-circTADA2A, which successfully inhibited the mRNA and protein expression of HMGN5 (Figure 3A-3C). To further explore the role of HMGN5 in circTADA2A-promoted proliferation, invasion, and migration of NSCLC cells, A549 and H1299 cells were transfected with si-HMGN5 or si-NC. si-HMGN5 blocked cell proliferation (Figure 3D,3E), migration (Figure 3F,3G), and invasion (Figure 3H) by silencing the mRNA expression of HMGN5. Most importantly, the correlation analysis confirmed a positive relationship between the circTADA2A and HMGN5 levels in human NSCLC tissues (Figure 3I). Taken together, these observations indicate that HMGN5 promotes NSCLC cell proliferation, invasion, and migration by upregulating HMGN5 expression.
CircTADA2A directly binds to miR-450b-3p
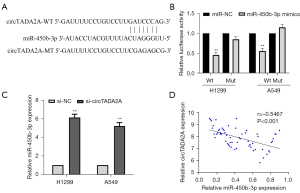
Based on the fact that circRNAs promote the progression of cancer by directly binding to miRNAs (17), we used circBank to predict the putative targeted miRNAs of circTADA2A to investigate whether circTADA2A could exert its function through miRNAs. This analysis identified miR-450b-3p as the miRNA that circTADA2A could target (Figure 4A). Next, dual-luciferase reporter analysis verified that miR-450b-3p mimics remarkably decreased the luciferase activity in circTADA2A-WT-transfected H1299 and A549 cells (Figure 4B). qRT-PCR analysis revealed that si-circTADA2A promoted the expression of miR-450b-3p in A549 and H1299 cells (Figure 4C). Furthermore, correlation analysis confirmed a negative relationship between circTADA2A and miR-450b-3p levels in human NSCLC tissues (Figure 4D). In summary, these results suggest that miR-450b-3p is the direct target of circTADA2A in NSCLC.
HMGN5 is the target of miR-450b-3p
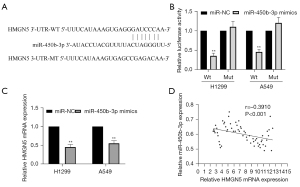
To further investigate the role of miR-450b-3p in the expression of HMGN5, we used StarBase software to predict the potential miR-450b-3p-binding sites in the 3'-UTR of HMGN5 (Figure 5A). A luciferase reporter assay confirmed that there was a binding interaction between miR-450b-3p and HMGN5 (Figure 5B). Additionally, miR-450b-3p mimics significantly inhibited the expression of HMGN5 in A549 and H1299 cells (Figure 5C), which was consistent with the negative correlation of miR-450b-3p and HMGN5 in human NSCLC tissues (Figure 5D). In conclusion, these findings demonstrate that HMGN5 is the target of miR-450b-3p in NSCLC.
Discussion
To date, increasing evidence has demonstrated that circRNAs lead to the occurrence and development of multiple types of cancers. NSCLC is the most common primary cancer in the pathophysiology of lung cancers. However, the functional role of circTADA2A in the pathogenesis of NSCLC and its underlying molecular mechanism remain unknown. In this research, we found that circTADA2A was markedly upregulated in human NSCLC tissues. The higher expression of circTADA2A was closely related to shorter overall survival of NSCLC patients. Moreover, circTADA2A promoted NSCLC cell proliferation, invasion, and migration by regulating HMGN5 expression. Notably, circTADA2A directly interacted with miR-450b-3p to upregulate HMGN5 expression.
CircTADA2A has been confirmed to accelerate several human cancers. Recently, it was reported that circTADA2A was significantly upregulated in human osteosarcoma tissues and advanced the progression of osteosarcoma (6). In addition, circTADA2A could also accelerate aerobic glycolysis in lung cancer cells by regulating the miR-455-3p/FOXM1 axis (9). However, the specific functional role of circTADA2A in the pathogenesis of NSCLC remains unclear. We also showed that circTADA2A served as a tumor enhancer in NSCLC, which was consistent with previous findings. Our results indicated that higher expression of circTADA2A was significantly associated with shorter overall survival of patients with NSCLC. In addition, circTADA2A promoted NSCLC cell proliferation, invasion, and migration. However, Li et al. reported that circMTO1 could act as a tumor suppressor in colorectal cancer by regulating the miR-374a-3p/KLF14 axis (7). Currently, the circRNAs including CircRNA_101237, circSATB2 and circP4HB promotes NSCLC (14,18,19) Conversely, circRNAs including circNDUFB2 and circPTPRA inhibits NSCLC progression (20,21). Our findings confirmed that circTADA2A promotes NSCLC proliferation which better improve researchers' understanding of lung cancer about circRNAs.
The functional roles and mechanisms of circRNAs in the occurrence and progression of cancer have not been fully clarified. Accumulating evidence has confirmed that circRNAs can regulate the expression of tumor-promoting or oncogenic genes by sponging miRNAs to further regulate the expression of tumorigenesis-correlated proteins (22-24). It was reported that circTADA2A promoted the progression and subsequent metastasis of osteosarcoma by sponging miR-203a-3p and upregulating CREB3 expression (6). Furthermore, circTADA2A could also regulate osteosarcoma proliferation by sponging miR-129-5p (25). HMGN5 is regarded as a tumor-enhancing gene due to its involvement in promoting the proliferation of various cancer cells (13,26,27), such as A549 and H1299 cells (28). In the present study, we demonstrated that circTADA2A directly interacted with miR-450b-3p and subsequently served as a miRNA sponge to upregulate HMGN5 expression. Although knockdown of circTADA2A inhibited the osteosarcoma progression and metastasis has been confirmed (6), the roles of circTADA2A in NSCLC in vivo need to be explored in future studies.
Currently, the clinical application of circRNA is often used as the early diagnostic biomarkers, treatment and prognosis evaluation. Due to the problems of medical ethics and challenges of circular conformation and sequence overlap with linear mRNA counterparts (29), the circRNA is unable to be applied in the medical field. In the future study, the well-designed circTADA2A interference drug development could be employed to treat NSCLC patients with the highly circTADA2A expression. In addition, the circTADA2A expression levels could be used to predict the prognosis of NSCLC patients.
Conclusions
We first confirmed that circTADA2A was significantly upregulated in NSCLC tissues and negatively associated with the prognosis of NSCLC patients. In addition, we also demonstrated that circTADA2A promoted the proliferation, invasion, and migration of NSCLC cells by sponging miR-450b-3p to elevate HMGN5 expression. From the perspective of translational medicine, these data verify that the circTADA2A/miR-450b-3p/HMGN5 axis could be a therapeutic target for the management of NSCLC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2836/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2836/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2836/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Zhuji People’s Hospital (No. 2021-0915). All patients provided written informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu G, Pei F, Yang F, et al. Role of Autophagy and Apoptosis in Non-Small-Cell Lung Cancer. Int J Mol Sci 2017;18. [Crossref] [PubMed]
- Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023-75. [Crossref] [PubMed]
- Garon EB, Hellmann MD, Rizvi NA, et al. Five-Year Overall Survival for Patients With Advanced NonSmall-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J Clin Oncol 2019;37:2518-27. [Crossref] [PubMed]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. [Crossref] [PubMed]
- Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci 2020;77:1661-80. [Crossref] [PubMed]
- Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer 2019;18:73. [Crossref] [PubMed]
- Li Z, Yao H, Wang S, et al. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res 2020;39:160. [Crossref] [PubMed]
- Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis 2019;10:175. [Crossref] [PubMed]
- Zhao H, Wei H, He J, et al. Propofol disrupts cell carcinogenesis and aerobic glycolysis by regulating circTADA2A/miR-455-3p/FOXM1 axis in lung cancer. Cell Cycle 2020;19:2538-52. [Crossref] [PubMed]
- Rochman M, Malicet C, Bustin M. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta 2010;1799:86-92. [Crossref] [PubMed]
- Wu J, Wang J. HMGN5 expression in bladder cancer tissue and its role on prognosis. Eur Rev Med Pharmacol Sci 2018;22:970-5. [PubMed]
- Yao K, He L, Gan Y, et al. HMGN5 promotes IL-6-induced epithelial-mesenchymal transition of bladder cancer by interacting with Hsp27. Aging (Albany NY) 2020;12:7282-98. [Crossref] [PubMed]
- Ma Q, Wang X, Wang H, et al. HMGN5 Silencing Suppresses Cell Biological Progression via AKT/MAPK Pathway in Human Glioblastoma Cells. Biomed Res Int 2020;2020:8610271. [Crossref] [PubMed]
- Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer 2020;19:101. [Crossref] [PubMed]
- Wang Y, Du J, Niu X, et al. MiR-130a-3p attenuates activation and induces apoptosis of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2. Cell Death Dis 2017;8:e2792. [Crossref] [PubMed]
- Zhao J, Wang Y, Wu X. HMGN5 promotes proliferation and invasion via the activation of Wnt/beta-catenin signaling pathway in pancreatic ductal adenocarcinoma. Oncol Lett 2018;16:4013-9. [PubMed]
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17:79. [Crossref] [PubMed]
- Zhang ZY, Gao XH, Ma MY, et al. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep 2020;10:9024. [Crossref] [PubMed]
- Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun 2019;513:904-11. [Crossref] [PubMed]
- Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine 2019;44:182-93. [Crossref] [PubMed]
- Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun 2021;12:295. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Bian A, Wang Y, Liu J, et al. Circular RNA Complement Factor H (CFH) Promotes Glioma Progression by Sponging miR-149 and Regulating AKT1. Med Sci Monit 2018;24:5704-12. [Crossref] [PubMed]
- Zhang J, Ma X, Zhou R, et al. TRPS1 and YAP1 Regulate Cell Proliferation and Drug Resistance of Osteosarcoma via Competitively Binding to the Target of circTADA2A - miR-129-5p. Onco Targets Ther 2020;13:12397-407. [Crossref] [PubMed]
- He J, Liu C, Wang B, et al. HMGN5 blockade by siRNA enhances apoptosis, suppresses invasion and increases chemosensitivity to temozolomide in meningiomas. Int J Oncol 2015;47:1503-11. [Crossref] [PubMed]
- Gan Y, Tan J, Yang J, et al. Knockdown of HMGN5 suppresses the viability and invasion of human urothelial bladder cancer 5637 cells in vitro and in vivo. Med Oncol 2015;32:136. [Crossref] [PubMed]
- Chen P, Wang XL, Ma ZS, et al. Knockdown of HMGN5 expression by RNA interference induces cell cycle arrest in human lung cancer cells. Asian Pac J Cancer Prev 2012;13:3223-8. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]


