
The cell cycle gene centromere protein K (CENPK) contributes to the malignant progression and prognosis of prostate cancer
Introduction
Prostate cancer (PCa) has become the second most common cancer in men, and both morbidity and mortality rates have increased in recent years (1). The occurrence of PCa may be affected by a heterogeneous set of factors, such as genes, cellular context, and environmental conditions (2,3). Existing biomarkers routinely used for PCa diagnosis and monitoring, including prostate-specific antigen (PSA), parametric magnetic resonance imaging (MRI), and hypermethylation of glutathione S-transferase pi gene (GSTP1) and prostate cancer gene 3 (PCA3) expression, have been identified in the past (4,5). However, the levels of sensitivity and specificity vary substantially, and additional biomarkers for PCa are needed to reduce overdiagnosis and overtreatment of the disease (6).
Androgen deprivation therapy (ADT) remains the gold standard for the treatment of advanced PCa, but the disease usually progresses to castration-resistant prostate cancer (CRPC) and is associated with a worse prognosis (7). While ADT is initially effective in most patients, it also promotes the expression of multiple genes that affect the cell cycle and mediate DNA replication, possibly contributing to the eventual emergence of CRPC (8,9). CENPK is a subunit of the centromeric complex and is required for proper kinetochore function and mitotic progression (10,11). The kinetochore, comprising at least 80 different proteins, is a protein complex assembled at each centromere, serving as the attachment site for spindle microtubules and the site at which motors generate forces to power chromosome movement (12). Dysregulation or dysfunction of kinetochores leads to aneuploidy and promotes carcinogenic effects (13). In addition, Okada et al. showed that CENPK conditional knockout cells stopped proliferating approximately 48 h after the addition of tetracycline, and most cells died by 96 h (14). Knockout and growth curve analyses revealed an important role for CENPK in cell proliferation. Studies have shown that CENPK is upregulated in ovarian cancer, triple-negative breast cancer, lung cancer and hepatocarcinoma (15-18). However, the clinical significance and mechanism of CENPK in PCa remain unclear.
In this study, we explored the function of CENPK in the pathogenesis of PCa. CENPK, a cell cycle-related gene, may be a potentially effective biomarker for PCa and contribute to malignant progression and prognosis. CENPK contributed to cell proliferation, colony formation, and tumorigenesis of CRPC in vitro. Gene enrichment and flow cytometry analyses have shown the involvement of the CENPK gene in mitosis in the cell cycle of cancer cells. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2164/rc).
Methods
Patient and tissue samples
Thirty-three primary PCa and 36 CRPC samples were collected from the Second Hospital of Tianjin Medical University. All primary PCa and CRPC tissues were confirmed by pathologists. Patients were diagnosed with CRPC according to the European Association of Urology (EAU) guidelines (19). All samples were collected from men who underwent radical prostatectomy (RP) or transurethral resection of the prostate (TURP) but not from any metastatic regions. Informed consent was obtained from the patients and their family members. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of The Second Hospital of Tianjin Medical University.
Bioinformatics analysis
The clinical information and expression profiles were obtained from The Cancer Genome Atlas (Prostate Adenocarcinoma, TCGA-PRAD) and Prostate Cancer Transcriptome Atlas (PCTA) datasets (20,21). The analysis was performed using the public prostate cancer microarray dataset (GSE8702) to determine changes in gene expression in LNCaP cells during 12 months of androgen deprivation (22). In this study, gene set enrichment analysis (GSEA) was used to examine whether CENPK plays an important role in the PCa cell cycle and contributes to malignant progression and prognosis in PCa using the R package clusterProfiler (version 3.6.2) (23). Gene set permutations were performed 1,000 times for each analysis using GSEA. An adjusted P value <0.05, false discovery rate (FDR) <0.25, and normalized enrichment score (|NES|) >1 were considered significant enrichment.
Immunohistochemistry (IHC)
Human and mouse prostate tissue specimens were cut into 4-µm sections and mounted on slides. PCa tissue sections were then deparaffinized, rehydrated and treated with antigen retrieval solution. Subsequently, sections were blocked with 5% FBS for 20 min. The sample sections were incubated with a primary antibody against CENPK (Bioss, Catalog: bs-8459R; Beijing, China) overnight at 4 ℃ and incubated with a secondary antibody at 37 ℃ for 1 h the next day. Finally, diaminobenzidine (DAB) was used to visualize the staining results.
Cell culture
LNCaP, LNCaP-AI, 22RV1, PC3 and DU145 cell lines were used in this study. LNCaP, 22RV1, PC3 and DU145 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). LNCaP-AI cells, a castration-resistant PCa cell line, were produced from LNCaP cells with long-term castration culture (24). LNCaP-AI cells were cultured in RPMI-1640 medium supplemented with 10% charcoal-stripped fetal bovine serum (CD-FBS) (BI, Cromwell, CT, USA) and a 1% penicillin streptomycin solution. LNCaP, 22RV1, PC3 and DU145 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (BI, Cromwell, CT, USA) and a 1% penicillin-streptomycin solution. All cells were incubated at 37 ℃ with 5% CO2.
Androgen stimulation and deprivation
For androgen stimulation, cells were starved for 72 h in RPMI 1640 media supplemented with 10% CD-FBS (BI, Cromwell, CT, USA) followed by stimulation with dihydrotestosterone (DHT) (Sigma-Aldrich, MO, USA) at the indicated time points. For long-term androgen deprivation, LNCaP cells were hormone-starved using RPMI 1640 media supplemented with 10% CD-FBS for up to 60 days, as described in a previous study (22).
Western blot
Cells were lysed with RIPA buffer containing 1% phenylmethylsulfonyl fluoride (PMSF) at 4 ℃ for 30 min. Protein samples were separated on a 10% SDS/PAGE gel and transferred onto a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was blocked with 5% nonfat milk for 1 h and incubated with the primary antibody at 4 ℃ overnight. After an incubation with a secondary antibody the next day, the membrane was exposed to enhanced chemiluminescence (ECL) reagent (Catalog: 32106; Thermo Scientific, MA, USA) to visualize bands. The primary antibodies used in this study were as follows: GAPDH (Abways, Catalog: AB0037; Shanghai, China) and CENPK (Bioss, Catalog: bs-8459R).
RNA interference
CENPK siRNAs and negative control siRNAs were purchased from Tianjin Sheweisi Co. The siRNAs and negative control siRNAs were transfected into LNCaP-AI and DU145 cells with GP-siRNA-Mate plus transfection reagent (GenePharma) according to the manufacturer’s protocol. The following siRNA sequences were used in this study: CENPK-si#1 target sequence: sense, 5'-CUCAGUCAAUGGCAGAAAATT-3' and antisense, 5'-UUUUCUGCCAUUGACUGAGTT-3'; and CENPK-si#2 target sequence: sense, 5'-GAAGACGUUCUCAUAACAUUATT-3' and antisense, 5'-UAAUGUUAUGAGAACGUCUUCTT-3'.
Quantitative real-time PCR
Total RNA was isolated from PCa cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The cDNA templates were synthesized using a reverse transcription kit (Roche). Real-time quantitative reverse transcription PCR (qRT-PCR) was performed with an Applied Biosystems 7900 Real-Time PCR System (Thermo Scientific). The primer sequences were as follows:
- GAPDH, forward 5'-CGGAGTCAACGGATTTGGTC-3' and reverse 5'-TCGCCCCACTTGATTTTGGA-3';
- CENPK, forward 5'-TTGTGACGCTGTGATGGTCT-3' and reverse 5’-ACGCTTGAGGATGCAAGATGT-3';
- KLK3, forward 5'-ACTGCATCAGGAACAAAAGCG-3' and reverse 5'-GAAGCTGTGGCTGACCTGAA-3';
- CCNB1, forward 5'-CGCCTGAGCCTATTTTGGTTG-3' and reverse 5'-AGTGACTTCCCGACCCAGTA-3';
- CCND1, forward 5'-GATCAAGTGTGACCCGGACTG-3' and reverse 5'-CCTTGGGGTCCATGTTCTGC-3';
- CCNE1, forward 5'-CCCATCATGCCGAGGGAG-3' and reverse 5'-TATTGTCCCAAGGCTGGCTC-3';
- CDKN1A, forward 5'-GCCGAAGTCAGTTCCTTGTG-3' and reverse 5'-TTCTGACATGGCGCCTCCT-3';
- CDKN2A, forward 5'-GGGGTCGGGTAGAGGAGG-3' and reverse 5'-GCCCATCATCATGACCTGGA-3';
- CDKN2B, forward 5'-TTTACGGCCAACGGTGGATT-3' and reverse 5'-CCATCATCATGACCT GGATCG-3';
- CDK1, forward 5'-CTTGGCTTCAAAGCTGGCTC-3' and reverse 5'-GGGTATGGTAGATCCCGGCT-3';
- CDK2, forward 5'-GATCAAGTGTGACCCGGACTG-3' and reverse 5'-CCTTGGGGTCCATGTTCTGC-3';
- CDK4, forward 5'-TGCGGCCTGTGTCTATGGTC-3' and reverse 5'-TCTCAGATCAAGGGAGACCCTCA-3'; and
- CDK6, forward 5'-TGCGGCCTGTGTCTATGGTC-3' and reverse 5'-TCTCAGATCAAGGGAGACCCTCA-3'.
MTT assay
CENPK-knockdown cells and NC cells were plated in 96-well plates (LNCaP-AI: 4×104 cells/well, DU145: 3×104 cells/well) at 37 ℃ with 5% CO2 for 1–6 days. Then, 100 µL of the MTT solution (5 mg/mL) were added to each well and incubated at 37 ℃ with 5% CO2 for 2 h. Finally, a microplate reader was used to measure the optical density (OD) value of each well at 490 nm. Each experiment was performed in triplicate.
Statistical analysis
Statistical significance was determined using either one-way ANOVA, two-way ANOVA with a post hoc multiple comparisons test, or unpaired two-tailed Student’s t-test for most experiments, unless indicated otherwise. P values <0.05 were considered significant: * represents P<0.05, ** represents P<0.01, *** represents P<0.001 and **** represents P<0.0001. Error bars represent standard deviations (SDs).
Results
Screening workflow of cell cycle-related genes in androgen-independent PCa
We created a bioinformatics-driven workflow to assess cell cycle-associated genes (Figure 1A). First, we used LNCaP cells, a castration-sensitive PCa cell line, as a model and evaluated molecular changes in gene expression during 12 months of androgen deprivation (GSE8702) (22). Next, we defined fast-response androgen-sensitive genes by comparing the genes that were dramatically downregulated (456 genes) in the first month of androgen deprivation culture compared to controls. Then, we compared the genes that were significantly upregulated at 5 months compared with 1 month (473 genes) and significantly upregulated at 12 months compared with 5 months (328 genes). Next, we constructed a Venn gram to screen genes that rapidly respond to androgen deprivation and determine the long-term emergence of androgen-independent overexpression patterns. Then, 14 genes were enriched in this workflow (Figure 1B). We screened cell cycle-related genes by selecting genes categorized into the cell cycle (biological process; GO: 0007049). Eleven of 14 genes were finally identified. Interestingly, BIRC5 (also known as Survivin) has been detected in prostate intraepithelial neoplasia from patients with both castration-sensitive and CRPC and is associated with a poor prognosis and an increased rate of recurrence (25). YM155, a small-molecule suppressor of BIRC5, has been tested in castration-resistant taxane-pretreated PCa (26). TOP2A, a key oncogene driving the growth of PCa cells, is overexpressed and associated with an increased risk of systemic progression in patients with PCa, and higher levels of TOP2A were also detected in patients with CRPC (27,28). These data reveal the robust nature of our screening process. Furthermore, we validated our findings by examining the expression patterns of 11 genes in the PCTA database (20). The 11 cell cycle-related gene signatures showed strong enrichment in the CRPC group, indicating higher expression in CRPC (Figure 1C,1D). We selected the CENPK gene to validate and discuss its clinical value in PCa, due to the lack of information regarding its clinical value of PCa.
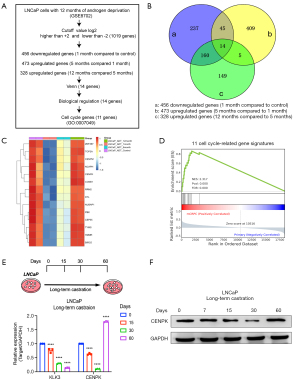
LNCaP cells were cultured under androgen deprivation conditions for 3 days followed by DHT treatment to validate the effect of androgen on CENPK expression. Along with KLK3, CENPK was upregulated by DHT treatment (Figure S1). LNCaP cells were cultured under androgen-deprived conditions for 60 days to estimate the effect of long-term androgen deprivation on CENPK expression (Figure 1E). Remarkably, in combination with prolonged androgen deprivation, a robust increase in CENPK expression was observed compared to KLK3 expression (Figure 1E,1F). Taken together, we successfully constructed a workflow to screen cell cycle-related genes in PCa and used CENPK as a target gene for further study.
Overexpression of CENPK in CRPC tissues
We assessed the CENPK expression pattern in the PCTA dataset, and the results showed an upregulation of CENPK expression in patients with CRPC compared to patients with primary PCa (Figure 2A). We further confirmed the upregulation of CENPK in CRPC tissues from our in-house dataset. Consistently, compared with patients with primary PCa, CENPK protein levels were evidently elevated in CRPC tissues, as determined using IHC (Figure 2B).
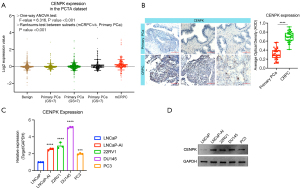
In addition, DNA methylation is the process of adding methyl groups to cytosine and adenine catalyzed DNA methyltransferase (29). When the level of promoter DNA methylation decreases, a process known as DNA demethylation, the inhibition of gene transcription is alleviated (30-32). We characterized the methylation status of the CENPK promoter in the PRAD dataset to preliminarily determine the mechanism of CENPK dysregulation (Figure S2). The results showed that CENPK promoter methylation levels were significantly reduced in primary PCa tissues, indicating that the upregulation of CENPK is associated with promoter methylation levels. Taken together, these data revealed that CENPK is expressed at higher levels in CRPC tissues than in primary PCa tissues, suggesting that CENPK may promote CRPC progression.
CENPK gene knockdown inhibits CRPC cell growth
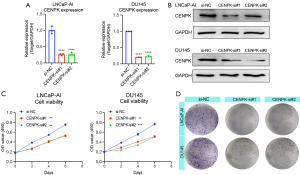
Next, we determined the CENPK expression profile in cell lines. Notably, all CRPC cell lines (LNCaP-AI, 22RV1, C4-2, DU145, and PC3) showed higher expression levels of CENPK than LNCaP cells (a castration-sensitive cell line), consistent with the expression pattern detected in patients. Among these cell lines, LNCaP-AI and DU145 cells showed relatively higher levels of CENPK in our study and were used for functional validation (Figure 2C,2D). We knocked down the endogenous expression of CENPK in LNCaP-AI and DU145 cells with two specific siRNAs to elucidate the possible biological functions of CENPK in CRPC cells (Figure 3A,3B). CENPK expression was prominently downregulated in both cell lines, as detected using qRT-PCR and Western blotting, which confirmed the knockdown efficacy. We studied the effect of CENPK silencing on proliferation by performing MTT and colony formation assays (Figure 3C,3D). The downregulation of CENPK significantly reduced cell viability and the number of colonies formed by LNCaP-AI and DU145 cells. Based on these data, CENPK plays a positive role in regulating CRPC cell growth.
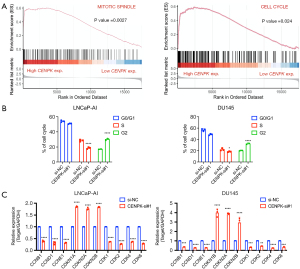
Pathway enrichment analysis reveals CENPK-related signaling pathways
We performed a pathway enrichment analysis using the PRAD dataset by dividing samples into a high CENPK expression group and a low CENPK expression group based on the median CENPK expression level to study the possible biological functions of CENPK (Figure S3). Gene sets enriched in the high CENPK expression groups were linked to mitotic spindles and the cell cycle, suggesting that CENPK may modulate the tumor-related signaling pathways that regulate cell proliferation and promote the development of CRPC (Figure 4A). Using a flow cytometry assay, we confirmed that dysregulated cell cycles were further disrupted by CENPK silencing (Figure 4B, Figure S4). In addition, the levels of the key regulatory factors of cell cycle progression, including cyclin B1 (CCNB1), cyclin D1 (CCND1), cyclin E1 (CCNE1), CDK2, and CDK4, were significantly decreased, while the levels of negative regulators of cell cycle progression, including CDKN1A (P21) and CDKN1B (P16), were increased in LNCaP-AI and DU145 cells compared with the controls, which was also consistent with the in vitro phenotype described in the present study (Figure 4C).
High CENPK expression represents a prognostic factor for patients with PCa
We constructed Kaplan-Meier curves to evaluate the relationship between CENPK expression in the PRAD dataset and survival (Figure 5A,5B). The median CENPK mRNA expression level was used as the cutoff point to split samples into the CENPK high and CENPK low groups. Patients with higher CENPK expression showed a lower probability of overall survival (OS) than the low CENPK group (P=0.031) and a lower probability of disease-free survival (DFS) (P=0.0081). In addition, patients exhibiting higher CENPK mRNA expression experienced shorter DFS and OS than the lower groups (Table 1). Therefore, patients with higher CENPK expression exhibit lower survival rates.
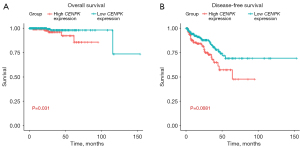
Table 1
| Characteristic | Total | The PRAD dataset* | ||
|---|---|---|---|---|
| High CENPK expression group | Low CENPK expression group | P value | ||
| Case, n [%] | 495 | 161 [32.5] | 334 [67.5] | – |
| Age, n [%] | 0.085 | |||
| ≤61 years | 249 | 72 [45] | 177 [53] | |
| >61 years | 246 | 89 [55] | 157 [47] | |
| Residual tumor, n [%] | 0.234 | |||
| Yes | 314 | 97 [64] | 217 [69] | |
| No | 151 | 55 [36] | 96 [31] | |
| Pathological stage, n [%] | 0.407 | |||
| < T3A | 187 | 56 [36] | 131 [40] | |
| ≥ T3A | 301 | 101 [64] | 200 [60] | |
| DFS, n [%] | 0.023 | |||
| Recurred/progressed | 70 | 31 [19] | 39 [12] | |
| Disease free | 425 | 130 [81] | 295 [88] | |
| OS, n [%] | <0.001 | |||
| Decease | 8 | 5 [3] | 3 [1] | |
| Alive | 487 | 156 [97] | 331 [99] | |
*, the PRAD dataset refers to the Prostate Adenocarcinoma (TCGA, TCGA Provisional) dataset; –, lack of relative information or no results. CENPK, centromere protein K; DFS, disease-free survival; OS, overall survival.
We performed univariate and multivariate Cox analyses of the PRAD dataset to further evaluate the clinical effect of CENPK on patients with PCa (Table 2). In the univariate analysis, high CENPK expression showed a significant correlation with a low DFS probability [hazard ratio (HR): 1.841, 95% confidence interval (CI): 1.148–2.951; P=0.011] and low OS probability (HR: 2.216, 95% CI: 1.127–4.009; P=0.020]. In the multivariate Cox analysis, high CENPK expression correlated with a low DFS probability (HR: 1.677, 95% CI: 1.030–2.732, P=0.038).
Table 2
| Variable | DFS | OS | |||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Univariate analysis | |||||
| Age | 1.026 (0.990–1.063) | 0.156 | 0.998 (0.896–1.111) | 0.969 | |
| Residual tumor | 1.600 (0.995–2.573) | 0.052 | 2.347 (0.520–10.600) | 0.267 | |
| Pathological stage | 0.764 (0.478–1.220) | 0.259 | 2.996 (0.581–15.450) | 0.190 | |
| CENPK expression | 1.841 (1.148–2.951) | 0.011 | 2.216 (1.127–4.009) | 0.020 | |
| Multivariate analysis | |||||
| Age | 1.018 (0.982–1.055) | 0.320 | 1.001 (0.905–1.107) | 0.991 | |
| Residual tumor | 1.474 (0.909–2.391) | 0.116 | 1.875 (0.361–9.751) | 0.455 | |
| Pathological stage | 0.764 (0.470–1.244) | 0.280 | 2.136 (0.383–11.930) | 0.387 | |
| CENPK expression | 1.677 (1.030–2.732) | 0.038 | 3.630 (0.683–19.300) | 0.130 | |
CI, confidence interval; CENPK, centromere protein K; DFS, disease-free survival; OS, overall survival.
Discussion
In our previous study, the expression of certain cell cycle-related genes was significantly upregulated in patients with PCa and potentially indicated shorter DFS and a poor prognosis (33). However, the study did not develop specific protocols or effective workflows to further characterize the functions of cell cycle-related genes in PCa. The goal of this study was to generate an experimentally proven bioinformatics workflow to screen cell cycle-related genes in PCa. Surprisingly, we found that CENPK might be used as a novel marker for patients with PCa and CRPC, and the results are promising. In this study, we characterized the differential expression patterns, potential biological functions, and mechanisms of CENPK in PCa.
CENPK is a protein contributing to the structure of the kinetochore on chromatids and is closely related to the regulation of chromosome segregation during mitosis and meiosis (12). The misregulation of centromeres contributes to chromosome mis-segregation and promotes carcinogenesis (13). Notably, the overexpression of many centromere genes appears to be strongly correlated with the malignant phenotype of cancer, although the mechanisms remain incompletely understood (34-36). In hepatocellular carcinoma (HCC), CENPK was identified as an oncogene that is upregulated by reducing promoter methylation and may contribute to carcinogenesis (37). CENPK overexpression is closely related to the clinicopathological characteristics of patients with HCC and was identified as an independent risk factor for the OS of patients. Moreover, Wang et al. found that CENPK overexpression stimulates the tyrosine phosphorylation of the Akt and MDM2 proteins and promotes cell proliferation while inhibiting tyrosine phosphorylation of the TP53 protein (17). According to Ma et al., CENPK significantly associates with CyclinB1, which triggers the transition from G2 phase to mitosis (18). In vitro, CENPK knockdown resulted in a significant reduction in cell proliferation, and flow cytometry analysis revealed that cell arrest and apoptotic capacity increased significantly following CENPK knockdown compared to the control group.
In this study, we clearly documented the CENPK expression level in multiple clinical tissues and multiple PCa cell lines, and preliminary analysis revealed a potential regulatory pathway of CENPK overexpression by reducing promoter methylation of CENPK in human PCa tissues. Consistently, Wang et al. also performed bisulfite DNA sequencing and showed that methylation of the CENPK promoter was markedly reduced in HCC specimens compared with adjacent noncancerous tissues (17). We performed a pathway enrichment analysis of CENPK-related genes, and the results revealed an association of CENPK overexpression with cell cycle pathways. In addition, during cell cycle analysis using flow cytometry, cells with CENPK silencing exhibited a prolonged G2 phase, consistent with disrupted cell proliferation. Taken together, our data suggest that CENPK knockdown inhibits CRPC cell proliferation and is a potential therapeutic target and diagnostic indicator.
However, our study has several limitations. For example, the expression of CENPK in androgen receptor (AR)-negative PCa cells (i.e., PC3 cells) remains to be investigated, and further studies are needed to clarify other regulatory mechanisms governing CENPK expression in PCa other than androgen-related mechanisms. In addition, the human samples used in our study were obtained from a single clinical center and represented a relatively small patient population. CENPK protein expression and activity must be clarified in larger patient samples and multiple clinical centers.
Acknowledgments
We thank AME Editing Service for English language editing.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2164/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2164/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2164/coif). The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of The Second Hospital of Tianjin Medical University (No. KY2021K016), and informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sekeroglu B, Tuncal K. Prediction of cancer incidence rates for the European continent using machine learning models. Health Informatics J 2021;27:1460458220983878. [Crossref] [PubMed]
- Haffner MC, Zwart W, Roudier MP, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol 2021;18:79-92. [Crossref] [PubMed]
- Saunders EJ, Kote-Jarai Z, Eeles RA. Identification of Germline Genetic Variants that Increase Prostate Cancer Risk and Influence Development of Aggressive Disease. Cancers (Basel) 2021;13:760. [Crossref] [PubMed]
- Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer 2008;8:268-78. [Crossref] [PubMed]
- Carroll PH, Mohler JL. NCCN Guidelines Updates: Prostate Cancer and Prostate Cancer Early Detection. J Natl Compr Canc Netw 2018;16:620-3. [Crossref] [PubMed]
- Lomas DJ, Ahmed HU. All change in the prostate cancer diagnostic pathway. Nat Rev Clin Oncol 2020;17:372-81. [Crossref] [PubMed]
- Jia S, Gao X, Lee SH, et al. Opposing effects of androgen deprivation and targeted therapy on prostate cancer prevention. Cancer Discov 2013;3:44-51. [Crossref] [PubMed]
- Gao S, Gao Y, He HH, et al. Androgen Receptor Tumor Suppressor Function Is Mediated by Recruitment of Retinoblastoma Protein. Cell Rep 2016;17:966-76. [Crossref] [PubMed]
- Pisano C, Tucci M, Di Stefano RF, et al. Interactions between androgen receptor signaling and other molecular pathways in prostate cancer progression: Current and future clinical implications. Crit Rev Oncol Hematol 2021;157:103185. [Crossref] [PubMed]
- Cheeseman IM, Hori T, Fukagawa T, et al. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell 2008;19:587-94. [Crossref] [PubMed]
- Hu L, Huang H, Hei M, et al. Structural analysis of fungal CENP-H/I/K homologs reveals a conserved assembly mechanism underlying proper chromosome alignment. Nucleic Acids Res 2019;47:468-79. [Crossref] [PubMed]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 2003;112:407-21. [Crossref] [PubMed]
- Scully R. The spindle-assembly checkpoint, aneuploidy, and gastrointestinal cancer. N Engl J Med 2010;363:2665-6. [Crossref] [PubMed]
- Okada M, Cheeseman IM, Hori T, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 2006;8:446-57. [Crossref] [PubMed]
- Komatsu M, Yoshimaru T, Matsuo T, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int J Oncol 2013;42:478-506. [Crossref] [PubMed]
- Lee YC, Huang CC, Lin DY, et al. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. PeerJ 2015;3:e1386. [Crossref] [PubMed]
- Wang H, Liu W, Liu L, et al. Overexpression of centromere protein K (CENP-K) gene in hepatocellular carcinoma promote cell proliferation by activating AKT/TP53 signal pathway. Oncotarget 2017;8:73994-4005. [Crossref] [PubMed]
- Ma J, Chen X, Lin M, et al. Bioinformatics analysis combined with experiments predicts CENPK as a potential prognostic factor for lung adenocarcinoma. Cancer Cell Int 2021;21:65. [Crossref] [PubMed]
- Heidenreich A, Bastian PJ, Bellmunt J, et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol 2014;65:467-79. [Crossref] [PubMed]
- Rotinen M, You S, Yang J, et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat Med 2018;24:1887-98. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011-25. [Crossref] [PubMed]
- D'Antonio JM, Ma C, Monzon FA, et al. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate 2008;68:698-714. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Shang Z, Yu J, Sun L, et al. LncRNA PCAT1 activates AKT and NF-κB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic Acids Res 2019;47:4211-25. [Crossref] [PubMed]
- Shariat SF, Lotan Y, Saboorian H, et al. Survivin expression is associated with features of biologically aggressive prostate carcinoma. Cancer 2004;100:751-7. [Crossref] [PubMed]
- Tolcher AW, Quinn DI, Ferrari A, et al. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann Oncol 2012;23:968-73. [Crossref] [PubMed]
- Schaefer-Klein JL, Murphy SJ, Johnson SH, et al. Topoisomerase 2 Alpha Cooperates with Androgen Receptor to Contribute to Prostate Cancer Progression. PLoS One 2015;10:e0142327. [Crossref] [PubMed]
- Labbé DP, Sweeney CJ, Brown M, et al. TOP2A and EZH2 Provide Early Detection of an Aggressive Prostate Cancer Subgroup. Clin Cancer Res 2017;23:7072-83. [Crossref] [PubMed]
- Lakshminarasimhan R, Liang G. The Role of DNA Methylation in Cancer. Adv Exp Med Biol 2016;945:151-72. [Crossref] [PubMed]
- Traube FR, Carell T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol 2017;14:1099-107. [Crossref] [PubMed]
- Lei Y, Huang YH, Goodell MA. DNA methylation and de-methylation using hybrid site-targeting proteins. Genome Biol 2018;19:187. [Crossref] [PubMed]
- Blanquart C, Linot C, Cartron PF, et al. Epigenetic Metalloenzymes. Curr Med Chem 2019;26:2748-85. [Crossref] [PubMed]
- Chen X, Ma Q, Liu Y, et al. Increased expression of CELSR3 indicates a poor prognostic factor for Prostate Cancer. J Cancer 2021;12:1115-24. [Crossref] [PubMed]
- El-Arabey AA, Salama SA, Abd-Allah AR. CENP-E as a target for cancer therapy: Where are we now? Life Sci 2018;208:192-200. [Crossref] [PubMed]
- Sharma AB, Dimitrov S, Hamiche A, et al. Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Res 2019;47:1051-69. [Crossref] [PubMed]
- Arunkumar G, Melters DP. Centromeric Transcription: A Conserved Swiss-Army Knife. Genes (Basel) 2020;11:911. [Crossref] [PubMed]
- Wang J, Li H, Xia C, et al. Downregulation of CENPK suppresses hepatocellular carcinoma malignant progression through regulating YAP1. Onco Targets Ther 2019;12:869-82. [Crossref] [PubMed]


