
Bioinformatics analysis reveals the clinical significance of GIPC2/GPD1L for colorectal cancer using TCGA database
Introduction
Colorectal cancer (CRC) is one of the most prevalent gastrointestinal malignancies and is the fourth most fatal tumor disease worldwide, causing approximately 900,000 deaths annually (1), generally accounting for 10% of all diagnosed cancer-related deaths globally each year (2). In addition to the aging population and eating habits in high-income countries, unfavorable causes like obesity, lack of physical exercise, smoking, and other adverse risk factors also increase the risk of CRC. Previous research in China aimed at assessing the factors influencing the known causes of cancer in 2005, including smoking, drinking, chronic infections, nutritional factors, overweight and obesity, physical inactivity, occupational factors as well as hormonal factors, which showed that nearly 14.6% of colon cancer deaths are due to alcohol consumption, overweight, obesity, and limited physical inactivity (3).
Although there are currently many influential diagnoses and treatments for CRC, it still poses a serious threat to millions of people worldwide. CRC is usually caused by mutations in oncogenes, tumor suppressor genes, and other specific genes, just as in other kinds of cancer (4); therefore, abnormal expression of many genes can affect the onset of CRC.
GIPC1, GIPC2, and GIPC3 consist of GIPC homology 1 (GH1) domain, PDZ domain, and GH2 domain. GIPC PDZ domain-containing family member 2 (GIPC2) is a protein encoded by GIPC2 gene in humans. GIPC2 has been reported as an endocrine-specific tumor suppressor gene for sporadic as well as hereditary tumors of RET- and SDHB- (5). Furthermore, GIPC2 is downregulated in acute lymphoblastic leukemia and upregulated in estrogen-induced breast tumors. Indeed, somatic mutations of GIPC2 occur in malignant melanoma and ovarian cancer (6) and are significantly linked to the survival of patients with papillary renal cell carcinoma (7). However, GIPC2 has not been reported in CRC.
GPD1L is a human gene. The protein encoded by this gene contains a glycerol-3-phosphate dehydrogenase (NAD+) motif, which shares 72% sequence homology with GPD1. miR-210-3p targets GPD1L to maintain HIF-1α stability, inhibit p53 activity through CYGB, and promote aerobic glycolysis (8). GPD1L negatively correlates with HIF1α expression and predicts lymph node metastasis of oral cavity and HPV-oropharyngeal carcinoma (9). Moreover, GPD1L is linked to occurrence and progression of head and neck squamous cell carcinoma, hepatocellular carcinoma, and breast cancer (10-12). However, the relationship between GPD1L and CRC is unclearly identified.
We used TCGA database to investigate the expression levels of GIPC2 and GPD1L in CRC and their correlation with clinical characteristics of CRC patients. In addition, we observed a strong association between GIPC2 and GPD1L expressions. Consequently, we analyzed signal pathways linked to GIPC2 and GPD1L. Our research provides that GIPC2 and GPD1L can be employed as diagnostic and prognostic markers for CRC, providing a theoretical basis for our subsequent cellular and animal experiments and is expected to further promote the level of CRC diagnosis and treatment. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1933/rc).
Methods
Cell culture
Human normal colon epithelial cells (HcoEpiC) along with CRC cell lines (Lovo, RKO, DLD-1, HCT116) were obtained from the Laboratory of Pathology at Dalian, Medical University, and then tested and authenticated. HcoEpiC, Lovo, and HCT116 cells were cultured with 10% fetal bovine serum (FBS) in Dulbecco’s Modified Eagle’s medium (DMEM) (GIBCO, USA). RKO and DLD-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) supplemented with 10% FBS. Incubations were performed at 37 ℃ and a 5% CO2 atmosphere.
Public data collection and GIPC2 protein expression in human CRC
Transcriptome RNA-seq data of CRC samples and relevant clinical information were obtained from TCGA database (https://portal.gdc.cancer.gov/). Meanwhile, clinical and pathological information (age, gender, tumor grade, stage, follow-up time, and survival status) was collected. TCGA databases and all cases involved in the database have been consented to be utilized for analyses and obtained ethical approval, which is free to be downloaded and analyzed by individual researchers. Our study was based on open-source data and strictly followed publication guidelines as well as access policies of the database. Accordingly, the study protocol was exempted from additional ethical approval.
GIPC2 protein expression images in normal tissues were determined from Human Protein Atlas (www.proteinatlas.org/ENSG00000137960-GIPC2/tissue/colon#img).
The CRC tissues images were determined from Human Protein Atlas (www.proteinatlas.org/ENSG00000137960-GIPC2/pathology/colorectal+cancer#img)
Correlation analysis
Pearson correlation analysis was applied to identify GIPC2- and GPD1L-related genes. Correlations were considered statistically significant when the absolute of Pearson correlation coefficient (|r|) >0.4 and P<0.05. GIPC2- and GPD1L-related gene expression patterns were visualized using R package of “pheatmap”. Subsequently, the Venn diagram was generated using R package “ggplot2” to determine the genes associated with GIPC2 and GPD1L expressions.
GO and KEGG enrichment analysis
Gene ontology (GO) was applied to identify the biological features of RNA-seq data. Separate gene ontology enrichment analysis using enrichment for upregulated and downregulated genes was performed. To determine which DEGs are activated and repressed in a different class of pathways, gene expression information was mapped to KEGG pathway. GO and KEGG enrichment analyses were performed for the genes correlated with GIPC2 and GPD1L expressions using R packages of clusterProfiler, enrichplot, and ggplot2. Terms with a P value <0.05 were considered significantly enriched.
Receiver operating characteristics (ROC) curve analysis and Survival Analysis
ROC curve analysis was applied using R package “pROC” to assess the diagnostic accuracy of GIPC2 and GPD1L. Kaplan-Meier (KM) survival analysis using R packages of “survminer” and “survival” was performed, and a log-rank test was applied to analyze the prognostic value of GIPC2 and GPD1L for overall survival (OS) and progress free interval (PFI) of CRC patients.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
Statistical analysis was performed in R environment (R version: 4.0.0). The Wilcoxon test (Mann-Whitney test) was used for continuous variable analysis. Fisher’s exact test or chi-square test was utilized for categorical data analysis. The difference in survival was analyzed using K-M analysis and log-rank test. For all statistical analyses, P<0.05 is considered statistically significant. The 2−ΔΔCt method was deployed to evaluate mRNA relative expression levels in PCR test.
Results
The association between GIPC2 expression and clinical characteristics
The role of GIPC2 in CRC has not yet been revealed; therefore, we first conjecture the relevance of GIPC2 with the clinical characteristics in CRC patients. In order to investigate this hypothesis, we analyzed the correlation between GIPC2 expression and clinicopathological staging factors, including T, N, M stage in CRC utilizing TCGA database. As demonstrated in Table 1, GIPC2 expression was closely correlated with a clinical-pathological stage (P<0.001). Additionally, TNM stage classification demonstrated a statistical significance in T and N stage (P=0.002, P<0.001, respectively); nevertheless, it did not reveal any significance in M stage (P=0.051), and no correlation was observed between GIPC2 expression and gender (P=0.647), as well as age (P=0.348).
Table 1
| Characteristic | Low expression of GIPC2 | High expression of GIPC2 | P value |
|---|---|---|---|
| N | 239 | 239 | |
| T stage, n (%) | 0.002* | ||
| T1 | 4 (0.8%) | 7 (1.5%) | |
| T2 | 30 (6.3%) | 53 (11.1%) | |
| T3 | 164 (34.4%) | 159 (33.3%) | |
| T4 | 41 (8.6%) | 19 (4%) | |
| N stage, n (%) | <0.001* | ||
| N0 | 122 (25.5%) | 162 (33.9%) | |
| N1 | 57 (11.9%) | 51 (10.7%) | |
| N2 | 60 (12.6%) | 26 (5.4%) | |
| M stage, n (%) | 0.051 | ||
| M0 | 168 (40.5%) | 181 (43.6%) | |
| M1 | 41 (9.9%) | 25 (6%) | |
| Pathologic stage, n (%) | <0.001* | ||
| Stage I | 29 (6.2%) | 52 (11.1%) | |
| Stage II | 86 (18.4%) | 101 (21.6%) | |
| Stage III | 79 (16.9%) | 54 (11.6%) | |
| Stage IV | 41 (8.8%) | 25 (5.4%) | |
| Gender, n (%) | 0.647 | ||
| Female | 116 (24.3%) | 110 (23%) | |
| Male | 123 (25.7%) | 129 (27%) | |
| Age, median (IQR) | 69 [58, 78] | 68 [59, 76] | 0.348 |
*, P<0.05 indicates a significant association between variables.
GIPC2 is downregulated in human CRC tissues and cells
To validate the function of GIPC2 in tumorigenesis, we searched CRC database of TCGA to assess the differential manifestation of GIPC2, which proves that cancer with GIPC2 transcripts (n=286) had suggestively lower manifestation level compared to normal samples (n=41) (Figure 1A). Moreover, to verify the manifestation of GIPC2 gene in CRC, we performed PCR assays with human normal colon epithelial cells (HcoEpiC) and CRC cell lines (Lovo, RKO, DLD-1, HCT116), which indicated the same results (Figure 1B). Moreover, we analyzed GIPC2 protein expression in clinical specimens through human protein atlas (www.proteinatlas.org) and found that GIPC2 was highly expressed in normal tissues compared with CRC tissues (Figure 1C). In brief, these findings demonstrated that GIPC2 is significantly downregulated in human CRC tissues and cells, both as mRNA and protein levels.
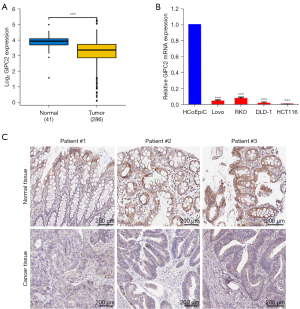
GIPC2 participates in CRC progression
Cancer analysis is a process that determines the degree of malignancy and spread of the primary tumor, most importantly, the degree of tumor invasion, lymph node metastasis, distant metastasis (TNM stage), and clinicopathologic stages are pivotal bases for clinicians to evaluate tumor progression and prognosis as well as therapeutic regimen formulation. Based on aforementioned results, we subsequently investigated GIPC2 impact on clinical stage and survival rate in CRC from TCGA database, clearly indicating that transcription levels of GIPC2 were significantly reduced in tissues with a higher degree of invasion in CRC patients (T stage) (Figure 2A, T1&T2 vs. T3&T4, **, P<0.01). In addition, it could be found in CRC patients that GIPC2 expression was decreased in tumor lymph node metastasis (N stage) (Figure 2B, N0 vs. N1&N2, ***, P<0.001) and was significantly suppressed in CRC tissues in patients with advanced clinicopathologic stages (Figure 2C, Stage I, II vs. Stage III, IV, ***, P<0.001). However, there is no statistical significance in distant tumor metastasis (M stage) (Figure 2D, M0 vs. M1, ns). These results suggested that GIPC2 may participate in the malignant progression of CRC.
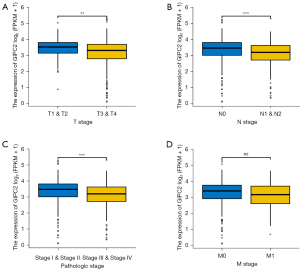
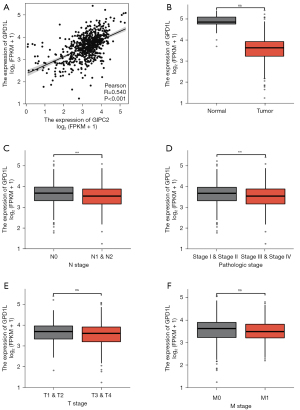
GPD1L is significantly correlated with GIPC2 expression and is essential for CRC progression
Tumor progression is induced by the interactions of multiple genes such as GIPC2, which has a significant role in CRC stage and is a vital gene influencing CRC occurrence and progression. We have revealed a significant correlation between GIPC2 and GPD1L expressions in CRC based on the correlation analysis (r=0.54, P<0.001) (Figure 3A). In addition, we verified in TCGA that GPD1L mRNA levels were significantly lower in CRC tumor tissues than normal tissues (Figure 3B, normal vs. tumor, ***, P<0.001). Furthermore, we examined the effect of GPD1L on the clinical stage of CRC, demonstrating that GPD1L expression was significantly inhibited in tumor lymph node metastasis (Figure 3C, N0 vs. N1&N2, **, P<0.01) and was significantly decreased in CRC tissues of patients with advanced clinicopathologic stages (Figure 3D, Stage I, II vs. Stage III, IV, **, P<0.01). However, there was no statistical significance in T and M stages (Figure 3E, T1&T2 vs. T3&T4, ns. Figure 3F, M0 vs. M1, ns). These results indicate that GPD1L is closely associated with decreased GIPC2 in CRC and is also involved in CRC progression.
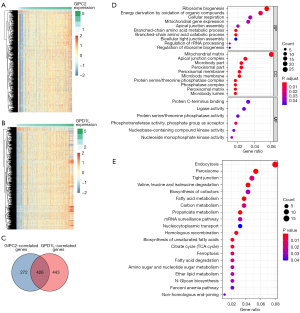
Functional exploration for GIPC2 and GPD1L in CRC
Regarding the potential significance of GIPC2 and GPD1L in CRC, we identified 698 GIPC2-related protein-coding genes (Figure 4A, online available: https://cdn.amegroups.cn/static/public/tcr-21-1933-01.xlsx) and 869 GPD1L-related protein-coding genes (Figure 4B, online available: https://cdn.amegroups.cn/static/public/tcr-21-1933-02.xlsx), respectively (r>0.4, P<0.05). By overlapping GIPC2- and GPD1L-related genes, 426 genes were acquired for subsequent GO and KEGG enrichment analyses (Figure 4C). The results of GO annotations, including biological process (BP), cellular components (CC), and molecular function (MF), are depicted in Figure 4D. In BP, the first three terms enriched for these 426 genes included ribosome biogenesis, energy derivation by oxidation of organic compounds, and cellular respiration. Regarding CC, the first three terms enriched included mitochondrial matrix, apical junction complex, and microbody part. Regarding MF, the first three terms enriched encompassed protein C-terminus binding, ligase activity, and protein serine/threonine phosphatase activity in MF. KEGG enrichment analysis revealed that these 426 genes are mainly enriched in endocytosis, peroxisome, tight junction, etc. (Figure 4E). We speculated that the roles of GIPC2 and GPD1L might be linked to the above enriched GO terms and KEGG pathways in CRC.
GIPC2 and GPD1L exhibited good diagnostic and prognostic predictive ability for CRC patients
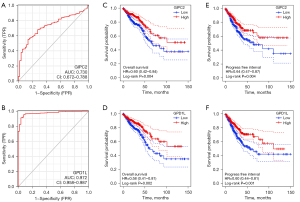
Furthermore, we analyzed whether GIPC2 and GPD1L expressions can be employed as diagnostic and prognostic markers for CRC patients using ROC curve analysis and KM survival analysis. Interestingly, ROC analysis revealed that the area under the curves (AUC) of GIPC2 and GPD1L were 0.73 and 0.972, respectively, implying that sensitivity and specificity of GIPC2 and GPD1L, especially GPD1L, are relatively satisfactory (Figure 5A,5B). To investigate the relationship between GIPC2/GPD1L expression and prognosis, CRC patients in TCGA database were divided into low and high groups according to their median values of GIPC2 or GPD1L. Subsequently, we conducted KM survival analysis and found that CRC patients in the low expression group of GIPC2/GPD1L demonstrated significantly shorter OS time and PFI than those in the high expression group (Figure 5C-5F). Therefore, we proposed that GPD1L and GIPC2 can be potential diagnostic as well as prognostic indicators for CRC patients.
Discussion
GIPC2 is a natural small molecule protein, which forms GIPC family together with GIPC1 and GIPC3. It consists of three domains: GH1, PDZ, and GH2. GIPC2 gene contains six exons and encodes 315 amino acids (5,6). GIPC2 gene is located in the homologous region of GIPC2 gene on the chromosome, and both are highly conserved in functional similarity. It is now known that GIPC2 expression has changed in various tumors, such as up-regulation in gastric cancer but down-regulation in kidney cancer, acute lymphoblastic leukemia, and adrenocortical carcinoma (5,7,13,14). However, the function of GIPC2 in the occurrence of colorectal tumors remains unclear. Studies have reported that GIPC2- associated genes are rarely used as tumor marker genes.
To determine the expression pattern of GIPC2 in CRC, we used TCGA data in colon cancer and qRT-PCR of cells, in which we discovered that compared with normal tissues and human normal colon epithelial cells, GIPC2 expression in CRC is significantly reduced. Additionally, we analyzed the correlation between GIPC2 expression and pathological features in CRC and stated that GIPC2 overexpression is intimately linked to CRC clinical-stage grade and TNM stage. These results revealed that GIPC2 may impact the early procedure during CRC initiation and progression and might exert a momentous function in CRC development.
Tumor occurrence and development occur due to multiple interactive factors. The protein encoded by glycerol triphosphate dehydrogenase 1-like antigen (GPD1L) is a protease that catalyzes the conversion of glycerol triphosphate to glycerol phosphate. The protein encoded by GPD1L is present in the cytoplasm associated with plasma membrane, where it binds to sodium channel voltage-gated V-type alpha subunit (SCN5a). The deficiency of this gene can induce Brugada syndrome type 2 (BRS2) and infants’ sudden death syndrome (SIDS) (15,16). Therefore, GIPC2 is critical for CRC, and we further investigated its correlated genes with GIPC2 using bioinformatics in CRC and disclosed that protein GPD1L is highly correlated with GIPC2 based on correlation analysis findings. Meanwhile, by analyzing the expression and clinical data of CRC patients and normal colorectal tissues from TCGA, we stated that GPD1L is distinctly downregulated in CRC compared to normal colorectal tissues. Furthermore, GPD1L overexpression is highly linked to CRC clinical-stage grade and TNM stage.
In addition, correlation analysis was applied, identifying 698 GIPC2- and 869 GPD1L-related genes. By overlapping GIPC2- and GPD1L-related genes, we obtained 426 genes, accounting for 61% GIPC2- and 49% GPD1L-related genes. These results further validated the close association between GIPC2 and GPD1L. We also investigated the underlying functions of these 426 genes in CRC through GO and KEGG analyses and revealed that GIPC2 and GPD1L were significantly correlated with the development of multiple tumorigenic-related signal pathways.
Moreover, ROC curve analysis and survival analysis using TCGA database in CRC indicated that the significant decrease in GPD1L and GIPC2 expressions might be early diagnostic and prognostic biomarkers. However, most current efforts in this area focused on narrowing the scope rather than enhancing the explanatory power of signature. In this study, the expressions of GIPC2 and GPD1L genes are clearly correlated, indicating orthogonality and functional complementarity between GIPC2 and GPD1L. As we hypothesized, GIPC2 and GPD1L demonstrated strong prognosis ability, suggesting the explanatory power of GIPC2/GPD1L marker, which played a fundamental role in CRC progression.
However, many limitations in our research did exist, we simply made some predictions and validation through some bioinformatics analysis and cell experiments. For the clinical aspects, we lacked the clinical research with our own patients tissue samples, in addition, further experiments in vivo and in vitro were absent either, which need to be improved in our follow-up research. But our results in this study were accurate, we obtained these results after extensive data analysis, which we believed provides a new research direction for the progress mechanism of CRC.
In summary, we proposed that GIPC2 and GPD1L are essential for CRC development and progression. GIPC2 and GPD1L expressions are significantly correlated, and they are associated with several critical cancer-related pathways, which might illustrate their underlying mechanisms in the incidence and CRC progression. Moreover, GIPC2 and GPD1L expressions not only have significant diagnosis potential but also demonstrate good prognosis value for CRC patients, providing a theoretical basis for our subsequent cellular and animal experiments. Through which, we hope to demonstrate the important function of GIPC2 in tumor progression, more importantly, we hope to provide a new tumor marker for diagnosis and prognosis of CRC, so as to reveal the occurrence and development mechanism of CRC more comprehensively.
Acknowledgments
We would like to thank El-Sayed Mohamed for his help in polishing our paper.
Funding: This work was supported by the Liaoning Science and Technology Project (No. 20180551172) and Dalian Medical Science Research Project (No. 1712044).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1933/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1933/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Wang JB, Jiang Y, Liang H, et al. Attributable causes of cancer in China. Ann Oncol 2012;23:2983-9. [Crossref] [PubMed]
- Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, et al. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci 2017;18:197. [Crossref] [PubMed]
- Dong Y, Huang Y, Fan C, et al. GIPC2 is an endocrine-specific tumor suppressor gene for both sporadic and hereditary tumors of RET- and SDHB-, but not VHL-associated clusters of pheochromocytoma/paraganglioma. Cell Death Dis 2021;12:444. [Crossref] [PubMed]
- Katoh M. Functional proteomics, human genetics and cancer biology of GIPC family members. Exp Mol Med 2013;45:e26. [Crossref] [PubMed]
- Liu Z, Wan Y, Yang M, et al. Identification of methylation-driven genes related to the prognosis of papillary renal cell carcinoma: a study based on The Cancer Genome Atlas. Cancer Cell Int 2020;20:235. [Crossref] [PubMed]
- Du Y, Wei N, Ma R, et al. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis 2020;11:731. [Crossref] [PubMed]
- Liu H, Wang S, Cheng A, et al. GPD1L is negatively associated with HIF1α expression and predicts lymph node metastasis in oral and HPV- Oropharyngeal cancer. Oral Dis 2021;27:1654-66. [Crossref] [PubMed]
- Feng Z, Li JN, Wang L, et al. The prognostic value of glycerol-3-phosphate dehydrogenase 1-like expression in head and neck squamous cell carcinoma. Histopathology 2014;64:348-55. [Crossref] [PubMed]
- Sulaiman SA, Abu N, Ab-Mutalib NS, et al. Signatures of gene expression, DNA methylation and microRNAs of hepatocellular carcinoma with vascular invasion. Future Oncol 2019;15:2603-17. [Crossref] [PubMed]
- Yoneten KK, Kasap M, Akpinar G, et al. Comparative Proteome Analysis of Breast Cancer Tissues Highlights the Importance of Glycerol-3-phosphate Dehydrogenase 1 and Monoacylglycerol Lipase in Breast Cancer Metabolism. Cancer Genomics Proteomics 2019;16:377-97. [Crossref] [PubMed]
- Kirikoshi H, Katoh M. Expression of human GIPC1 in normal tissues, cancer cell lines, and primary tumors. Int J Mol Med 2002;9:509-13. [Crossref] [PubMed]
- Kirikoshi H, Katoh M. Up-regulation of GIPC2 in human gastric cancer. Int J Oncol 2002;20:1183-7. [Crossref] [PubMed]
- Cui XF, Zhang SL, Wang WP, et al. Identification of competing endogenous RNA network in laryngeal squamous cell carcinoma. Oral Dis 2021; [Epub ahead of print]. [Crossref] [PubMed]
- Tu K, Li J, Mo H, et al. Identification and validation of redox-immune based prognostic signature for hepatocellular carcinoma. Int J Med Sci 2021;18:2030-41. [Crossref] [PubMed]


