
Expression, purification, and kinetic characterization of the human strep-IDO1
Introduction
L-tryptophan (L-Trp) is an essential amino acid acquired from food intake, and over 90% of it degraded through the kynurenine (Kyn) pathway (KP) as the substrate for indoleamine-2,3-dioxygenase 1 (IDO1) (1). IDO1 is generally expressed in various tissues and cells, ensuring the immune system’s balance in healthy human body (2-5). In the tissue microenvironment, overexpression of IDO1 leads to depletion of Trp, which further inhibits T cell proliferation and induces T cell exhaustion and apoptosis (6). Growing evidences have shown that IDO1 mediates immunosuppression and neovascularization in the tumor microenvironment, suggesting that it can be used as a novel target for tumor immunotherapy (7,8). Given the vital role in tumor immune escape, IDO1 is one of the most studied targets of cancer immunotherapies (9). Various IDO1 inhibitors with different structure have been developed, such as epacadostat (INCB024360), indoximod (D-1MT), navoximod (NLG919), BMS-986205 (Ono-7701/linrodostat), PF-06840003 (EOS-200,271), KHK-2455, LY-3381916 and DN1406131 (10,11). However, the disappointing results of phase III clinical trial of IDO1 inhibitor (epacadostat) combination with anti-PD1 antibody (pembrolizumab) in patients with malignant melanoma highlight the urgent need to further develop more effective IDO1 inhibitor (12).
The purification of IDO1 with high purity is one of the major constraints that limit the development of its inhibitors (13). Tag-assisted protein expression and purification is a powerful tool for producing recombinant proteins in academic research and large-scale industrial demands (14). An appropriate tagging strategy must provide sufficient yield and high purity for the final protein products while preserving their native structure and function (15). His, glutathione S-transferase (GST), maltose-binding protein (MBP), and FLAG were commonly used tags in previous study. MBP and GST are well-known large tags that can interfere with the structure and function of the main protein (14). Thus, they must be removed via the enzyme at the subsequent purification process. The expression and purification of IDO1 in E. coli based on His-tag have been reported previously (16-18). His-tag IDO1 can be purified from E. coli lysate using immobilized microparticles decorated with Ni, Zi or Co chelators such as nitrilotriacetic acid (NTA) (19). Further purification work is needed because of the modest specificity of His-tag, causing a low purity product. So far, using a minimal process to produces high purity recombinant IDO1 remains challenging. Therefore, it is necessary to develop an effective method to purify large quantities of IDO1 with high purity for further pharmacological research.
In the present study, we report a rapid one-step purification protocol for IDO1-strep recombinant protein with improved purity. This purification strategy of IDO1 presented here can be applied to multiple fields of cancer research investigating immune escape mechanism, whilst also contributing to the development of effective inhibitors. We also prepare polyclonal antibodies against IDO1 as a tool for further research on the function of this protein.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2518/rc).
Methods
Primer design
The forward IDO1-F and reverse IDO1-R primers for PCR amplification and P1, P2, P3 primers for sequencing were synthesized by Beijing Tianyi Huiyuan Life Science & Technology Inc. (Beijing, China). The primer sequences were as follows: 5'-ATGGGTCGCGGATCCGAATTCATGGCACACGCTATGGAAAACT-3' as IDO1-F and 5'-GTGGTGGTGGTGGTGCTCGAGTTTTTCGAACTGAGGGTGAGACCAACCTTCCTTCAAAAGGGATTTC-3' as IDO1-R. The primer sequences for sequencing were as follows: 5'-TAATACGACTCACTATAGG -3' as P1; 5'-AAAGGATTCTTC CTGGTCTCTCTATT-3' as P2 and 5'-CCCCAAGGGGTTATGCTAG-3' as P3.
Animal and cell line
Eight-week-old C57BL/6 mice, all female, were purchased from the Center of Medical Experimental Animals of the Chinese Academy of Medical Science. These animals were maintained in a sterile environment under a 12 h light-dark cycle and with free access to food and water. Animal experiments were performed following the guidelines of the Chinese Council on Animal Care. The research protocol was approved by the Animal Care and Use Committee at Chinese Academy of Medical Science. Human breast cancer cell line MCF-7 was obtained from the Cell Resource Centre of Peking Union Medical College and cultured in DMEM medium (Gibco) with 10% FBS (Gibco). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Plasmid and reagents
The Human IDO cDNA ORF Clone plasmid and pET-28a were purchased from Sino Biological Inc. (Beijing, China), Escherichia coli transetta (DE3) and trans5α competent cells were obtained from TransGen Biotech Corporation (Beijing, China). L-Kyn was from Solarbio (Beijing, China). Sodium acetate and glacial acetic acid were purchased from Sinopharm Chemical Reagent Co. (Shanghai, China). IFN-γ was purchased from R&D Systems (MN, USA). Other reagents were provided by Beyotime Biotechnology (Shanghai, China).
Plasmid construction
The Human IDO cDNA ORF Clone plasmid was used as a template, and the IDO1-F and IDO1-R primers were used to PCR amplify the IDO1 gene. The reaction condition was 10 µL 5× GC Buffer, 4 µL dNTP Mixture, 32.5 µL H2O, 0.5 µL PrimeStar HS DNA polymerase (TaKaRa, Japan). Reaction parameters were as follows: predenaturation at 98 ℃ for 5 min, 30 cycles of denaturation at 98 ℃ for 10 s, annealing at 55 ℃ for 5 s, and extension at 72 ℃ for 1min. After the reaction was completed, all of the PCR product was electrophoresed in a 1% agarose gel, and the target fragment at 1,200 bp was recycled by gel. The pET-28a plasmid was digested with EcoR I and Xho I restriction enzymes (Thermo Fisher Scientific, USA), recovered and purified, and ligated with PCR amplification product by using 5× In-Fusion HD Enzyme (TaKaRa, Japan) at 50 ℃ for 15 min. Extracting the recombinant plasmid from positive clones and double digested with EcoR I and Xho I restriction enzymes for identification and sent to Beijing Tianyi Huiyuan Life Science & Technology Inc. for sequencing by using primer P1, P2 and P3. The correct sequence recombinant plasmid was named pET28a-IDO1-strep.
Expression of recombinant IDO1
To test the optimal expression conditions of IDO1, single colonies of E. coli Transetta (DE3) transformed with the recombinant plasmid pET28a-IDO1-strep were inoculated in the tube containing 20 mL of LB medium, supplemented with 50 mg/mL of kanamycin (Solarbio, Beijing), and cultivated at 37 ℃ in a shaking incubator until the OD600nm of the culture was about 0.4. The expression of IDO1 was induced by a group of isopropyl β-D-thiogalactoside (IPTG) (Solarbio, Beijing) at varied concentrations containing 0.05, 0.1, 0.5, and 1.0 mmol/L, with or without 0.5 mM 5-Aminolevulinic acid hydrochloride (5-ALA) (Solarbio, Beijing). After induction for 24 h at 16 ℃, the cell was harvested and disrupted by sonication. Then the cell lysate was electrophoresed in 10% polyacrylamide gels and analyzed by a silver staining kit (Beyotime, Shanghai).
Purification of recombinant IDO1
After induced with 0.05 mM IPTG and 0.5 mM 5-ALA for 24 h at 16 ℃, 1 L of culture cells transformed with the recombinant plasmid pET28a-IDO1-strep were harvested by centrifugation at 6,000 g for 10 min at 25 ℃. The cell pellets were lysed by sonication in 10 mM Tris-HCl (pH =7.4), 150 mM NaCl, 1 mM EDTA, 1 mg/mL lysozyme, and protease inhibitor (Solarbio, Beijing). The supernatant was added to Magrose Strep-Tactin beads (beaverbio, Suzhou) after being obtained by centrifuged at 20,000 rpm for 1 h at 4 ℃. Then the mixture was incubated at 4 ℃ in a rotary shaker for 4 h. After incubation, the wash buffer (10 mM Tris-HCl, pH =7.4, 150 mM NaCl, 1 mM EDTA, pH=8.0) removed certain impurities. The Strep-tagged proteins were eluted with the same buffer containing 2.5 mM desthiobiotin (Sigma, USA). The beads were washed 3 times and eluted 3 times, and all solutions were collected, then electrophoresed in 10% polyacrylamide gels and analyzed by Coomassie Blue R-250 staining kit (Beyotime, Shanghai). The purified protein was dialyzed by Slide-A-Lyzer Dialysis Cassettes (Thermo Fisher Scientific, USA) in preservation buffer (20 mM Tris-HCl, pH=7.4) and 150 mM NaCl) 24 h at 4 ℃. Protein concentration was assessed in the supernatants by BCA Protein Assay Kit (Beyotime, Shanghai). The purified protein was further analyzed by SDS-PAGE and Western blot.
IDO1 enzyme activity
Two µg purified protein was added into 200 µL of the reaction buffer consists of 20 mM ascorbic acid sodium (pH =6.5), 50 mM sodium phosphate, 800 nM bovine liver catalase, 10 µM methylene blue (Solarbio, Beijing), and L-Trp (Sigma-Aldrich, USA). The mixture was incubated for 5–60 min at 37 ℃ and stopped by adding 50 µL trichloroacetic acid (2 mM) (Sigma-Aldrich, USA). After incubation at 65 ℃ for 15 min, the mixture was centrifuged at 15,000 g for 10 min at 4 ℃. The Kyn in the supernatant was analyzed by high performance liquid chromatography (HPLC).
To measure the enzymatic parameters, Michaelis-Menten constants (Km) were determined using different concentrations of L-Trp and the reaction time was 5 min. The initial velocity was measured within 5 min reaction time. Kinetic parameters (Km, Vmax) were determined from Lineweaver-Burk plots. All data were analyzed using GraphPad Prism Version 8 (GraphPad Software, Inc., La Jolla, CA, USA).
The UltiMate 3000 analytical HPLC system with UltiMate 3000 Autosampler with column compartment (Thermo Fisher Scientific, USA) was used for the measurements. Chromatography was carried out at 30 ℃. The HPLC system was equipped with an analytical column (XBridge® C18 3.5 µm; 4.6 mm × 250 mm column) was controlled by Chromeleon software. The chromatographic separation was achieved with a mobile phase of 15 mM sodium acetate in water (pH adjusted to 4.0±0.05 with glacial acetic acid) and 8% acetonitrile (Thermo Fisher Scientific, USA). The mobile phase was filtered through a 0.45 µm membrane filter and degassed by ultrasonic equipment for 15 min before use. The flow rate was 1.1 mL/min, and the injection volume was 100 µL. The retention time of Kyn (monitored at 360 nm by UV detector) is about 5.6 min. The peak factions at 5.6 min were collected for Mass spectra analysis.
Polyclonal antibody preparation
Ten mice were divided into two groups (immunization group and negative control group) according to the random number table method, five in each group. Immunization group mice were first immunized subcutaneously using 50 µg of recombinant protein thoroughly mixed in Freund’s complete adjuvant (Sigma-Aldrich, USA). The immunization intervals were 0, 7, 14, and 28 days, and an incomplete Freund adjuvant (Sigma-Aldrich, USA) was used for subsequent immunization. Seven days after the last immunization, mice were anaesthetized and serum was collected by extracting the eyeball.
Western blot
MCF-7 cells were treated with interferon-γ (10 ng/mL) or PBS for 24 h first. According to the user guide, the total proteins isolated from pET28a and MCF-7 cells or the purified protein were collected and determined by BCA Protein Assay Kit (Beyotime, Shanghai). Then, the protein was run on an SDS-PAGE gel and transferred to nitrocellulose membranes. The membranes were blocked in QuickBlock™ Western Blocking Buffer (Beyotime, Shanghai) for 30 min at room temperature, then were incubated with anti-IDO1 antibody (Cell Signaling, Cat.: 86630S; 1:1,000), anti-6× His antibody (Cell Signaling, Cat.: 12698S; 1:1,000), GAPDH (Cell Signaling, Cat.: 5174S; 1:1,000), or antiserum (1:200 dilution) overnight at 4 ℃. The next day, after the primary antibody was washed out using TBST, the nitrocellulose membranes were further incubated with peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Proteintech, USA; 1:2,000) for 1 h at room temperature. Finally, the target proteins were visualized using SuperSignal™ West Atto Substrate Solution (Thermo Fisher Scientific, USA).
Immunofluorescence
MCF-7 cells were treated with interferon-γ (10 ng/mL) for 24 h, then fixed in 4% paraformaldehyde for 10 min, permeabilized with Triton X-100 (Beyotime, Shanghai), and blocked with Immunol Staining Blocking Solution (Beyotime, Shanghai) for 30 min at room temperature, then were incubated with antiserum or negative serum (1:100 dilution) overnight at 4 ℃. The next day, the cells were washed three times in PBS and subsequently incubated with Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Thermo Fisher Scientific, USA; 1:1,000) for 1 h at room temperature. Cell nuclei were stained with the DAPI solution (1 µg/mL). Images were obtained using a confocal microscope (Nikon, Japan).
ELISA
96-well multiple well plates (Corning, USA) were coated with 100 µL purified protein (50 µg/mL), incubated overnight at 4 ℃. The wells were washed with PBST and blocked with 100 µL of 5% BSA for 1 h at room temperature. Then the wells were incubated with 100 µL antisera against IDO1 and negative sera with different deliquations (1:100, 1:500, 1:1,000, 1:2,000, 1:4,000, 1:8,000, 1:16,000, 1:32,000, 1:64,000) at 37 ℃ for 2 h. After the antiserum were washed out, the wells were incubated with 100 µL horseradish peroxidase-conjugated goat anti-mouse IgG (Proteintech, USA) for 2 h at 37 °C. A total of 100 µL TMB Chromogen Solution for enzyme linked immunosorbent assay (ELISA) (Beyotime, Shanghai) was added to each well, incubated at room temperature for 15 min; then the reaction was stopped by 100 µL Stop Solution for TMB Substrate (Beyotime, Shanghai). The absorbance was measured at 450 nm by the multi-mode reader (Biotek, USA).
Mass spectrum analysis
Mass spectrum analysis was performed on a Triple-TOF 5600 mass spectrometer (AB Sciex). The MS data were acquired using high-sensitivity mode with the following parameters: DDA mode, full scans acquired at a resolution of 40,000 and MS/MS scans at a resolution of 20,000; heater temperature, 550 ℃; Gas 1,50; Gas2, 35; spray voltage, 4.5 kV, Collision energy: 35 for the positive mode. A full scan and MS/MS scan range ranges from 50 to 1,250 (m/z) was used. Raw data files were processed using Progenesis QI (Waters, Milford, MA, USA) software. Compound identification was performed by searching the HMDB database (2019 version) and metlin MS/MS library using an MS1 tolerance of 10 ppm and an MS2 tolerance of 20 ppm.
Statistical analysis
In expression level analysis, we downloaded the standardized pan-cancer data (PANCAN, N=19,131, G=60,499) from the Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/), Genotype-Tissue Expression Project (GTEx) (https://www.genome.gov/), Therapeutically Applicable Research to Generate Effective Treatments (TARGET) (https://ocg.cancer.gov/programs/target) using UCSC Xena (https://xenabrowser.net/) and extracted the expression data of ENSG00000131203 (IDO1) gene in each sample. Then, we performed log2(x+0.001) transformation for each expression value and eliminated the cancer samples with less than three samples in a single cancer species. Finally, we obtained the expression data of 44 cancer species and used R software (version 3.6.4) to calculate the expression difference between normal and tumor samples. Unpaired Wilcoxon Rank Sum Test and Signed Rank Test were used for significance analysis of differences.
In survival analysis, we used the maxstat R package to calculate the optimal cut-off value of ENSG00000131203 (IDO1). The minimum number of samples was set to be greater than 25%, and the maximum number of samples was set to be less than 75%. Based on the optimal cut-off value (−4.6082), the patients were divided into high and low groups. We used the survival R package to establish Cox proportional hazard regression model (20) to analyze the relationship between gene expression and prognosis in each tumor. Survival curves were constructed using the Kaplan-Meier method, and the differences between the survival curves were examined by log rank test. The Survival R package was further used to analyze the difference in prognosis between the two groups.
To explore the relationship between IDO1 expression and immunity, the ESTIMATE algorithm was used to calculate ESTIMATE, immune and stromal scores for each tumor sample by utilizing estimate R package. The relationship between IDO1 expression and these three scores was evaluated using psych R packages. Correlations were examined with Pearson’s correlation test.
Results
Significance of IDO1 expression in tumor immunology
To analyze the expression level of IDO1 gene in different tumors and corresponding normal tissues, we performed a comprehensive analysis using TCGA, TARGET, GTEx databases. In comparison with normal tissue, IDO1 was significantly overexpressed in most solid tumors, including the brain, breast, cervical, ovarian, uterine, and endometrial cancer (Figure 1A). We then explored the prognostic value of IDO1 by using TCGA and TARGET database and obtained the expression data of 44 cancer species and the overall survival data of the corresponding samples. Furthermore, Kaplan-Meier analysis suggested that high expression of IDO1 jeopardized the overall survival of kidney renal papillary cell carcinoma (KIRP), brain lower-grade glioma (LGG), uveal melanoma (UVM) patients (Figure 1B). Therefore, IDO1 may serves as an indispensable material in tumor progression. Estimation of Stromal and Immune Cells in Malignant Tumor Tissues Using Expression Data (ESTIMATE) is a method for inferring the degree of stromal or immune cells infiltration into tumors using existing gene expression profiles (21). Moreover, to investigate relationship between IDO1 expression the tumor microenvironment, we obtained the immune infiltration scores of 10180 tumor samples in 44 tumor types. Our data implied that the expression of IDO1 was positively correlated with immune infiltration in 40 cancer species. The three tumors with the highest correlation coefficients are presented in Figure 1C. These results implicated that IDO1 plays a crucial role in tumor immunology.
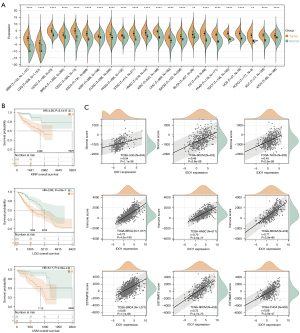
Construction of pET28a-IDO1-strep
The recombinant IDO1 protein can be purified to explore the detailed mechanisms. Using Human IDO cDNA ORF Clone as a template and IDO1-F and IDO1-R as primers for PCR amplification, 1% agarose gel electrophoresis showed the IDO-strep DNA fragment with a size of approximately 1,200 bp was amplified (Figure 2A). Then the DNA fragment was recovered and ligated with the linearized pET-28a vector (Figure 2B), which digested by EcoR I and Xho I, using In-Fusion enzyme. The recombinant plasmid was digested by the restriction enzymes EcoR I and Xho I. Agarose gel electrophoresis of the digest revealed two DNA bands at about 1,200 and 5,000 bp, which is equal to the theoretically calculated value of the expected length of the IDO1 gene fragment and digested pET-28a plasmid respectively (Figure 2C). The inserted fragment was verified by sequencing, which was identical to the IDO1 gene sequence in GeneBank. The recombinant resulting plasmid of 6574bp was named pET28a-IDO1-strep, and its DNA map is shown below (Figure 2D).
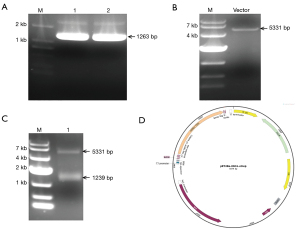
Optimizations of expression conditions and protein purification
IDO1 is a heme-dependent enzyme, the addition of the natural biosynthetic precursor of the heme (protoporphyrin IX), δ-Aminolevulinic acid, increased heme content of other hemoproteins, and guaranteed activation of the recombinant protein (17). To explore the optimum conditions that produce high yields of the recombinant IDO1, a suitable E. coli strain competent cells, Rosetta (DE3), were transformed with the pET28a-IDO1-strep plasmid and induced with isopropyl β-D-thiogalactoside (IPTG) at four different concentrations (0.05, 0.1, 0.5, and 1.0 mM) and with or without 5-Aminolevulinic acid hydrochloride (5-ALA) (0.5 mM). By comparing the intensity of the protein band after the sample was subjected to SDS-PAGE and silver staining, the expression level of the protein induced with different IPTG concentrations can be assessed (Figure 3A,3B). As the results show, 0.05 mM IPTG and 0.5 mM 5-ALA were finally selected as the optimum induced conditions.
The protein is purified by strep-tactin beads within a rapid one-step purification. A single band corresponding to the recombinant IDO1 protein was observed in a Coomassie blue stained SDS-PAGE gel after elution with a buffer adding 2.5 mM desthiobiotin (Figure 3C). The molecular weight of the protein product was approximately 50 kDa. Western blot analysis using an anti-His antibody and anti-IDO1 antibody confirmed that the purified protein is indeed IDO1 (Figure 3D). Based on the bicinchoninic acid assay measured protein concentration after dialysis, the final yield of the purified IDO1 was approximately 5 mg/L of Luria-Bertani (LB) culture medium.
Enzymatic activity analysis and identification of polyclonal antibody against IDO1
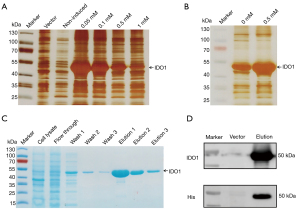
The IDO1 activity analysis was based on measuring Kyn generation by HPLC (22). To initiate an enzymatic reaction, we add the recombinant protein IDO1 to a reaction buffer consists of ascorbic acid sodium (reducing agent), methylene blue (reducing agent), bovine liver catalase (decomposing the hydrogen peroxide produced by the reducing agent), and L-Trp as the enzyme-substrate (23,24). As shown in Figure 4A, Kyn was produced only recombinant IDO1 protein and L-Trp were present in the reaction buffer, and the catalytic reaction product Kyn was confirmed by mass spectrometry (Figure 4B). After demonstrating the catalytic activity of recombinant IDO1 protein, a Michaelis-Menten curve obtained for this enzyme is shown in Figure 4C,4D, the Km value was 0.3015 mM and kcat value was 36.5 min−1. In addition, the initial velocity measured within 5 min reaction time was 6.027 nmol/min (Figure 4E).
Since IDO is a heme enzyme, we want to detected the content of holo-enzyme and heme of recombinant IDO1 protein. As shown in Figure 4F, the Abs404/Abs280 ratio of recombinant enzyme was 0.743 based on the UV-visible spectrum. Additionally, the amount of the holo-enzyme was 0.24 mg/mL according to the absorbance of the Soret peak using an extinction coefficient.
After immunizing mice with recombinant IDO1 protein as described in material and methods, anti-IDO1 serum was collected and determined by ELISA and the titer was approximately 1:16,000 (Figure 4G). At the same time, normal mice serum used as negative control did not result in a detectable signal. Western blot analysis showed that the antibody recognizes recombinant IDO1 protein and endogenous IDO1 from interferon-γ treated MCF-7 cell lysate (Figure 4H,4I). Immunofluorescence staining results demonstrate that the newly prepared polyclonal antibody can be used in detecting the endogenous IDO1 (Figure 4J).
Discussion
Immunotherapy has emerged as a standard pillar of cancer treatment in addition to surgery, radiotherapy, chemotherapy, and molecular-targeted therapy (25). Instead of directly killing cancer cells, cancer immunotherapy mediates the interaction between tumor microenvironment and immune cells or evaluates the specificity of cancer cell antigen by modifying immune cells to express an artificial receptor (26). Unfortunately, a considerable number of patients cannot benefit from immunotherapy due to the tumoral immune evasion (27). Nowadays, combination therapy with targeted drugs have been developed and discovered to overcome tumor-induced immune suppression (9). Inhibitor drug discovery often begins with the screening of a high number of chemical compounds against a therapeutic protein target in vitro (28). Consequently, it is urgent to explore a feasible and low-cost method to purify IDO1 with high purity.
Our research showed enhanced IDO1 expression in 22 types of cancer according to TCGA, TARGET, and GTEx database. Kaplan-Meier survival analysis using TCGA data demonstrated that high IDO1 expression was linked to dismal prognosis in KIRP, LGG, and UVM. According to ESTIMATE scores, IDO1 expression positively correlated with both stromal and immune cell content in the tumor microenvironment of 40 types of cancer. These results hinted that expression of IDO1 is closely related to immune infiltration of tumor cells. In conclusion, we found IDO1 was a key target for developing inhibitors and selected it as the target for further expression and purification. Using a purified protein to elucidate mechanisms of disease is of importance in biomedical research. Although there is a wide variety of recombinant protein commercially available, not all proteins have significant activity. Among the several kinds of methods for protein purification, E.coli expression system is often the first choice due to its easy operation, short time for expression, and low equipment requirements (29). In this study, recombinant IDO1 protein was highly efficiently expressed in E. coli strain competent cells Rosetta (DE3) using the pET28a expression vector. The final yield of the protein was approximately 5 mg/L of LB by using one-step purification through strep-tactin beads. The Enzymatic activity and kinetic parameters of the IDO1 obtained using HPLC. The Km value was 0.3015 mM and the kcat value was 36.5 min−1. The polyclonal antibody raised against the protein could specifically recognize IDO1.
Previous studies have reported the expression and purification of his-IDO1 in E. coli and their enzymatic properties, including substrate specificity (16-18,30,31). However, the purification of protein using Ni-NTA-agarose leads to inferior purity, and a size exclusion chromatography column is essential for further purification. In this work, strep-tag was utilized to satisfy our requirement of producing highly purified protein within a rapid one-step purification. The Strep-tag II is a protein tag of an eight-residue peptide sequence (WSHPQFEK) with an intrinsic affinity to streptavidin (32). IDO1 expressed by pET28a-IDO1-strep plasmid was flanked with a Strep-tag at the C-terminus, which can highly specifically bind to immobilized Strep-Tactin beads. The particular binding of strep-tag to strep-tactin and mild elution buffer conditions guarantees the protein’s high purity (32). To our knowledge, this study has practical application and it reports the purification of IDO1 based on strep-tag for the first time. This work lays the groundwork for future investigations on IDO1 protein function and provides systematic experimental methods for similar studies. As an effective method, this technique could be applied to a variety of proteins that are difficult to express and purify in further studies. This finding may expand our outlook on potential anti-IDO1 drug screening.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2518/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2518/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2518/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Animal experiments were performed following the guidelines of the Chinese Council on Animal Care. The research protocol was approved by the Animal Care and Use Committee at Chinese Academy of Medical Science.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Takikawa O, Yoshida R, Kido R, et al. Tryptophan degradation in mice initiated by indoleamine 2,3-dioxygenase. J Biol Chem 1986;261:3648-53. [Crossref] [PubMed]
- Thomas SR, Salahifar H, Mashima R, et al. Antioxidants inhibit indoleamine 2,3-dioxygenase in IFN-gamma-activated human macrophages: posttranslational regulation by pyrrolidine dithiocarbamate. J Immunol 2001;166:6332-40. [Crossref] [PubMed]
- Zhai L, Spranger S, Binder DC, et al. Molecular Pathways: Targeting IDO1 and Other Tryptophan Dioxygenases for Cancer Immunotherapy. Clin Cancer Res 2015;21:5427-33. [Crossref] [PubMed]
- Dolšak A, Gobec S, Sova M. Indoleamine and tryptophan 2,3-dioxygenases as important future therapeutic targets. Pharmacol Ther 2021;221:107746. [Crossref] [PubMed]
- Platten M, Nollen EAA, Röhrig UF, et al. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 2019;18:379-401. [Crossref] [PubMed]
- Lemos H, Huang L, Prendergast GC, et al. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat Rev Cancer 2019;19:162-75. [Crossref] [PubMed]
- Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017;357:eaaf9794. [Crossref] [PubMed]
- Cheong JE, Sun L. Targeting the IDO1/TDO2-KYN-AhR Pathway for Cancer Immunotherapy - Challenges and Opportunities. Trends Pharmacol Sci 2018;39:307-25. [Crossref] [PubMed]
- Xin Yu J, Hubbard-Lucey VM, Tang J. Immuno-oncology drug development goes global. Nat Rev Drug Discov 2019;18:899-900. [Crossref] [PubMed]
- Le Naour J, Galluzzi L, Zitvogel L, et al. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology 2020;9:1777625. [Crossref] [PubMed]
- Liu XH, Zhai XY. Role of tryptophan metabolism in cancers and therapeutic implications. Biochimie 2021;182:131-9. [Crossref] [PubMed]
- Yao Y, Liang H, Fang X, et al. What is the prospect of indoleamine 2,3-dioxygenase 1 inhibition in cancer? Extrapolation from the past. J Exp Clin Cancer Res 2021;40:60. [Crossref] [PubMed]
- Muheem A, Shakeel F, Jahangir MA, et al. A review on the strategies for oral delivery of proteins and peptides and their clinical perspectives. Saudi Pharm J 2016;24:413-28. [Crossref] [PubMed]
- Mahmoudi Gomari M, Saraygord-Afshari N, Farsimadan M, et al. Opportunities and challenges of the tag-assisted protein purification techniques: Applications in the pharmaceutical industry. Biotechnol Adv 2020;45:107653. [Crossref] [PubMed]
- Ki MR, Pack SP. Fusion tags to enhance heterologous protein expression. Appl Microbiol Biotechnol 2020;104:2411-25. [Crossref] [PubMed]
- Littlejohn TK, Takikawa O, Skylas D, et al. Expression and purification of recombinant human indoleamine 2, 3-dioxygenase. Protein Expr Purif 2000;19:22-9. [Crossref] [PubMed]
- Austin CJ, Mizdrak J, Matin A, et al. Optimised expression and purification of recombinant human indoleamine 2,3-dioxygenase. Protein Expr Purif 2004;37:392-8. [Crossref] [PubMed]
- Röhrig UF, Reynaud A, Majjigapu SR, et al. Inhibition Mechanisms of Indoleamine 2,3-Dioxygenase 1 (IDO1). J Med Chem 2019;62:8784-95. [Crossref] [PubMed]
- Shao S, Geng J, Ah Yi H, et al. Functionalization of cobalt porphyrin-phospholipid bilayers with his-tagged ligands and antigens. Nat Chem 2015;7:438-46. [Crossref] [PubMed]
- Andersen PK, Gill RD. Cox’s Regression Model for Counting Processes: A Large Sample Study. The Annals of Statistics 1982;10:1100-20. [Crossref]
- Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One 2017;12:e0179726. [Crossref] [PubMed]
- Zhao J, Gao P, Zhu D. Optimization of Zn2+-containing mobile phase for simultaneous determination of kynurenine, kynurenic acid and tryptophan in human plasma by high performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci 2010;878:603-8. [Crossref] [PubMed]
- Sadok I, Rachwał K, Jonik I, et al. Reliable chromatographic assay for measuring of indoleamine 2,3-dioxygenase 1 (IDO1) activity in human cancer cells. J Enzyme Inhib Med Chem 2021;36:581-92. [Crossref] [PubMed]
- Amirkhani A, Heldin E, Markides KE, et al. Quantitation of tryptophan, kynurenine and kynurenic acid in human plasma by capillary liquid chromatography-electrospray ionization tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2002;780:381-7. [Crossref] [PubMed]
- Ma Q, Chen Y, Xiao F, et al. A signature of estimate-stromal-immune score-based genes associated with the prognosis of lung adenocarcinoma. Transl Lung Cancer Res 2021;10:1484-500. [Crossref] [PubMed]
- Finn OJ. Immuno-oncology: understanding the function and dysfunction of the immune system in cancer. Ann Oncol 2012;23 Suppl 8:viii6-9.
- Sui M, Li Y, Wang H, et al. Two cases of intrahepatic cholangiocellular carcinoma with high insertion-deletion ratios that achieved a complete response following chemotherapy combined with PD-1 blockade. J Immunother Cancer 2019;7:125. [Crossref] [PubMed]
- Wu J, Liu B, Chan WKB, et al. Precise modelling and interpretation of bioactivities of ligands targeting G protein-coupled receptors. Bioinformatics 2019;35:i324-32. [Crossref] [PubMed]
- Kamionka M. Engineering of therapeutic proteins production in Escherichia coli. Curr Pharm Biotechnol 2011;12:268-74. [Crossref] [PubMed]
- Meininger D, Zalameda L, Liu Y, et al. Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and discovery of selective IDO1 inhibitors. Biochim Biophys Acta 2011;1814:1947-54. [Crossref] [PubMed]
- Luo S, Xu K, Xiang S, et al. High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form. Acta Crystallogr F Struct Biol Commun 2018;74:717-24. [Crossref] [PubMed]
- Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc 2007;2:1528-35. [Crossref] [PubMed]


