
Identification of a three-miRNA panel in serum for bladder cancer diagnosis by a diagnostic test
Introduction
Bladder cancer (BC) is one of the most common urinary malignant tumors and is the tenth most common cancer around the world (1). There would be about five hundred and forty-nine thousand new cases of BC and two hundred thousand deaths affected by BC a year (1). However, the 5-year survival rate of BC is 77%, it is closely related to tumor type, stage, and grade. The 5-year survival rate sharply reduces to 35% when the BC cells spread to the surrounding tissues and the 5-year survival rate is no more than less 10% when migrating into distant organs (2). Thus, the early detection of BC without metastasis is the key to improving the survival rate of patients. The gold standard for BC detection is the cystoscopy, an invasive and costly method. It gives a huge burden to the patient family and the national healthcare system. This has greatly increased the cost of treatment and surveillance of BC (3). Also, the cystoscopy would increase the possibility of the urinary tract infection, even for flexible cystoscopy (4). Therefore, it is necessary to find novel feasible diagnostic methods for BC.
In recent years, the research on diagnostic biomarkers of BC has aroused great interest, but most of them have not been applied clinically (5,6). There are currently 6 urinary assays, including NMP22 enzyme-linked immunosorbent assay (ELISA), NMP22 BladderChek, UroVysion, immunocyte (UCyt+), BTA-TRAK, and BTA-STAT, approved by the US Food and Drug Administration to enhance diagnosis and surveillance of BC. However, these tests are still not sensitive enough, and false positives usually occur in benign conditions, leading to urinary tract inflammation (7). The new, non-invasive and reliable diagnostic biomarkers are urgently needed for detecting BC.
MicroRNAs (miRNAs) are non-coding and endogenous approximately 22 nucleotides RNAs that can play important regulatory roles through binding the 3' untranslated region of protein-coding genes, leading to the degradation of messenger RNAs or destruction of the protein translation process (8,9). Numerous studies in recent years have reported that miRNAs could be secreted from cells into body fluids, including urine, blood, tears and gastric juice and they are highly stable by being packaged in exosome particles from the degradation of RNase. Then, circulating miRNAs have become reliable and noninvasive biomarkers for reflecting certain diseases, including tumors (10-12). Also, the prospect of miRNAs in the diagnosis and prognosis of urinary tumors is exciting and miRNAs have great potential as biological fluid indicators of urinary system tumors, such as urine and serum (13). A 7-miRNA panel was identified by Usuba et al. to be a serum biomarker for the early and specific screening of BC with the highest accuracy (AUC: 0.97; sensitivity: 95%; specificity: 87%) (2). Thus, these evidences make serum miRNA perform as a novel diagnostic biomarker for BC possible.
Here, a three-stage study was conducted to verify potential circulating serum biomarkers for BC by the profile of miRNAs expression in serum based on quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Then the diagnosis values of key miRNAs were analyzed for the detection of BC. Furthermore, bioinformatics analyses were used for exploring more biological functions related to targeted genes of the key miRNAs. We present the following article in accordance with the STARD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2611/rc).
Methods
Participants and ethics statement
Including 137 BC patients and 127 healthy controls (HCs), a total of 264 participants were recruited for serum samples between June 2017 and October 2020 from Peking University Shenzhen Hospital. The patients were clinically diagnosed with BC by cystoscopy and cystoscopic biopsies and confirmed by surgical biopsies. The HCs were conducting the routine physical examination in Peking University Shenzhen Hospital and without the abnormal laboratory findings and history of cancer. The characteristics of the participants are exhibited in Table 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Peking University Shenzhen Hospital approved all study protocols (approval ID. 2017-007). All participants recruited in our study agreed to use their serum samples for research purposes and provided written informed consent.
Table 1
| Variables | Screening stage (n=40) | Training stage (n=60) | Validation stage (n=164) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BC | HC | P value | BC | HC | P value | BC | HC | P value | |||
| Total number | 25 | 15 | 30 | 30 | 82 | 82 | |||||
| Age years, mean ± SD | 62.2±12.6 | 64.9±5.5 | 0.45 | 63.7±13.5 | 65.5 ±13.2 | 0.60 | 61.2±13.5 | 62.7±9.1 | 0.41 | ||
| Gender, n (%) | 0.87 | 0.61 | 0.76 | ||||||||
| Male | 14 (56.0) | 8 (53.3) | 18 (60.0) | 16 (53.3) | 42 (51.2) | 44 (53.7) | |||||
| Female | 11 (44.0) | 7 (46.7) | 12 (56.7) | 14 (46.7) | 40 (47.8) | 38 (46.3) | |||||
| Histological grade, n (%) | |||||||||||
| Low | 12 (48.0) | 20 (66.7) | 43 (52.4) | ||||||||
| High | 13 (52.0) | 10 (33.3) | 39 (47.6) | ||||||||
Among three stages, there was no significant difference between BC and HCs in age and gender. Parameters were shown as number (percentage). Statistical contrast was exerted through the Kruskal-Wallis rank test. BC, bladder cancer; HC, healthy control.
Study procedure
First, the 28 miRNAs related to BC were selected as candidate biomarkers for the next study from the Gene Expression Omnibus database or literature on the PubMed, following the retrieval strategies as (“Urinary Bladder Neoplasms”[Mesh] OR (bladder cancer [Title/Abstract])) AND (miRNA OR microRNA OR “MicroRNAs”[Mesh])). Then, as shown in Figure S1, a three-stage study was conducted to verify the candidate biomarkers. Five BC pools and three HC pools were set up at the screening stage. We randomly selected 25 serum samples from BC patients and 15 serum samples from HCs and combined every five samples into one pool. The expression profile of 28 miRNAs was detected by high throughput qRT-PCR, and we found differentially expressed miRNAs between two groups. Next, in the training stage, differently expressed miRNAs were confirmed by 30 serum samples from BC patients and 30 serum samples from HCs by qRT-PCR. Then, 82 BC patient samples and 82 HCs samples were utilized to evaluate the expression difference of final key miRNAs by qRT-PCR in the validation stage. Last, a miRNAs compound panel was constructed for verifying the diagnostic value.
Sample collection and RNA extraction
Before initial treatment, 10 mL of venous blood sample has been taken from participants. Within 2 hours after collection, the whole blood was separated into cell parts and serum at 4 ℃ and by centrifugation at 1,000 g for 10 minutes and then, at 15,000 g for 5 minutes. Before the process of extraction and purification, 2 µL of synthetic C. elegans miR-54-5p (cel-miR-54-5p) (10 nM/L, RiboBio, Guangzhou, China) was spiked into each serum sample to control variability. Total RNA from serum was extracted under the guidance of operation instruction of TRIzol LS isolation kit (Thermo Fisher Scientific, Waltham, MA, USA) and was added 30 µl RNase-free water for dissolution. Then they were kept at −80 ℃ for further processing. The purity of RNA was evaluated by NanoDrop 2000 spectrophotometer (NanoDrop, Wilmington, DE, USA).
qRT-PCR
The specific primers of reverse transcription from Bulge-Loop miRNA qRT-PCR Primer Set (RiboBio) were used to amplify miRNAs. The amplification and detection of miRNAs were conducted in 384-well plates on LightCycler 480 Real-Time PCR System (Roche Diagnostics, Mannheim, Germany), using the Taqman probe. The procedures were at 95 ℃ for 5 minutes, followed by 35 cycles of 95 ℃ for 10 s, 60 ℃ for 30 s. The expression of miRNAs in serum was calculated by the 2−∆∆Cq method and normalized to exogenous reference miRNA cel-miR-54-5p (14).
The Cancer Genome Atlas (TCGA) dataset analysis and bioinformatic analysis
Kaplan-Meier survival analysis and the log-rank test were conducted by OncoLnc (http://www.oncolnc.org/) to investigate the overall survival rate of BC patients, which links TCGA survival data with miR-132-3p, miR-7-5p and miR-148b-3p expression levels (15). To further study the functional involvement of the key miRNAs in BC, miRWalk3.0 (http://mirwalk.umm.uni-heidelberg.de/) was used for targeted genes prediction and the Enrichr database (amp.pharm.mssm.edu/Enrichr/) was used for Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. And GO include molecular function (MF), biological process (BP), and cellular component (CC) (16).
Statistical analysis
The information between groups on demographic and clinical characteristics was expressed as percentages or counts, while continuous variables were exhibited as means ± standard deviation. The Kruskal-Wallis rank test was conducted multiple comparisons among separate independent phases. The combined diagnostic test and the miRNA signature was conducted by the multiple logistic regression analysis with the expression profile of key miRNAs in the validation stage. The diagnostic value of the key miRNAs for BC were assessed by receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC). The optimal sensitivity and specificity and optimum cutoff values were determined by the Youden index (calculated as J = Sensitivity + Specificity − 1). Our statistical analyses were conducted by GraphPad Prism 8 (GraphPadSoftware Inc, La Jolla, CA, USA), SPSS software (SPSS 20.0 Inc, Chicago, IL, USA) and Medcalc (Version 19). We defined the P value <0.05 in two-sided as statistical significance.
Results
Characteristics of study participants
Including 137 BC patients and 127 HCs, a totally of 264 participants were involved in this study. The patients were confirmed by surgical biopsies based on World Health Organization standards, who diagnosed clinically with BC. These HCs matched BC patients based on age and gender, without history of cancer or other benign diseases. Table 1 showed the clinical and demographics characteristics of these participants in our study. No apparent difference in the distribution of gender or age has been found between the BC patients and HCs, with P values >0.05.
Candidate miRNAs in the screening stage
In the screening stage, the expression level of 28 miRNAs was screened out in 5 BC pools and 3 HCs pools to distinguish the candidate miRNAs (Figure 1). The candidate miRNAs were selected according to the differential expression (1.5-fold change and P<0.05) between BC pools and HCs pools. As a result, the 10 miRNAs, including six down-regulated significantly miRNAs and four up-regulated significantly miRNAs, were chose as candidate miRNAs to study in the next stage.
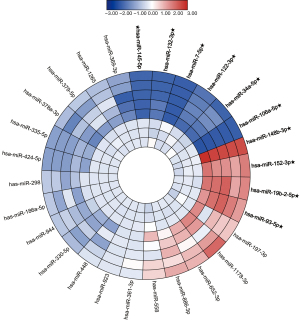
Further confirmation of candidate miRNAs in the training stage
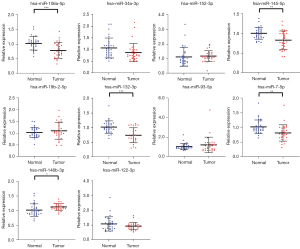
With 30 BC patients and 30 HCs, the screened 10 candidate miRNAs selected from the screening stage were assessed by qRT-PCR analysis in the training stage. The five miRNAs (miR-106a-5p, miR-145-5p, miR-132-3p, miR-7-5p, and miR-148b-3p), among the 14 candidate miRNAs, were still significantly expressed differentially in the serum between BC patients and HCs, as shown in Figure 2. Therefore, these five miRNAs were selected for study in the validation stage.
Expression level and diagnostic value of candidate miRNAs in the validation stage
The expression level of five key serum miRNAs was assessed with 82 BC patients and 82 HCs. The five candidate miRNAs were still significantly dysregulated between BC patients compared with HCs, as shown in Figure 3. And the expression of miR-148b-3p was increased significantly in BC patients, while the other four miRNAs, including miR-106a-5p, miR-145-5p, miR-132-3p, and miR-7-5p showed a contrary result.
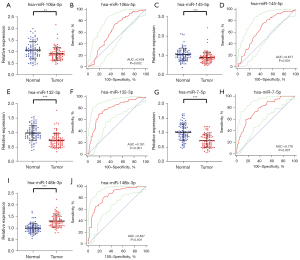
The diagnostic value of the five candidate miRNAs was evaluated by ROC curves analysis. Respectively, the AUCs were 0.634 [95% confidence interval (CI): 0.555 to 0.707; Figure 3B], 0.677 (95% CI: 0.600 to 0.748; Figure 3D), 0.781 (95% CI: 0.710 to 0.842; Figure 3F), 0.778 (95% CI: 0.707 to 0.839; Figure 3H) and 0.837 (95% CI: 0.771 to 0.890; Figure 3J) for miR-106a-5p, miR-145-5p, miR-132-3p, miR-7-5p and miR-148b-3p, as shown in Table 2. In addition, Table 2 listed the best sensitivity and specificity of these candidate miRNAs in diagnosing BC and the optimum cutoff values calculated by Youden index.
Table 2
| Variables | AUC | P value | 95% CI | Associated criterion | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| miR-106a-5p | 0.634 | 0.0025 | 0.555 to 0.707 | ≤1 | 73.17 | 53.66 |
| miR-145-5p | 0.677 | <0.001 | 0.600 to 0.748 | ≤0.98 | 75.61 | 56.10 |
| miR-132-3p | 0.781 | <0.001 | 0.710 to 0.842 | ≤0.83 | 68.29 | 81.71 |
| miR-7-5p | 0.778 | <0.001 | 0.707 to 0.839 | ≤0.76 | 59.76 | 84.15 |
| miR-148b-3p | 0.837 | <0.001 | 0.771 to 0.890 | >1.1 | 81.71 | 71.95 |
| Three-miRNA panel | 0.922 | <0.001 | 0.870 to 0.958 | >0.42286 | 90.24 | 81.71 |
miRNA, microRNA; AUC, area under curve; CI, confidence interval.
MiRNA panels construction for detection of BC better
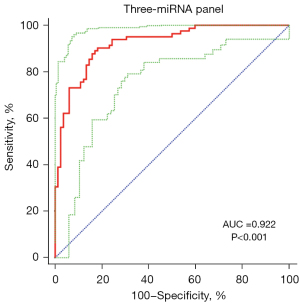
To diagnose BC better, a combination of several miRNAs could improve the accuracy in diagnosis than one single miRNA. Based on the stepwise logistic regression model, the expression profile of five key miRNAs in the validation stage was combined to set up diagnostic panels. Consequently, a three-miRNA panel including miR-132-3p, miR-7-5p, and miR-148b-3p was building for diagnosis of BC, and the model was calculated with the formula: Logit(P) =−0.918 − 2.641 × miR-132-3p − 4.880 × miR-7-5p + 6.581 × miR-148b-3p. The AUC for the three-miRNA panel was 0.922 (95% CI: 0.870 to 0.958; sensitivity =90.24%, specificity =81.71%; Figure 4).
Bioinformatics analysis and survival analysis of candidate miRNAs
The miRWalk 3.0 was used for target genes prediction of candidate miRNAs (miR-132-3p, miR-7-5p and miR-148b-3p). As shown in Figure 5, a totally of 366 genes were selected as target genes, predicted in more than two miRNAs. Through GEPIA, the expression of the 23 genes predicted in all candidate miRNAs in BC patients was determined by data from TCGA and the GTEx projects (17). With |log2FC| cutoff >1.5, P<0.01, BNC2, GAS7, and NTRK2, significantly differentially expressed in BC patients compared to HCs, were considered as potential target genes of the three-miRNA panel.
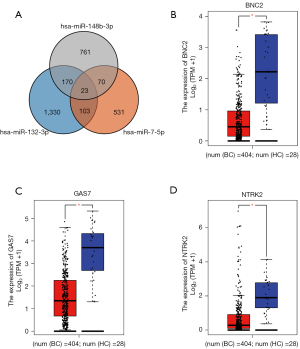
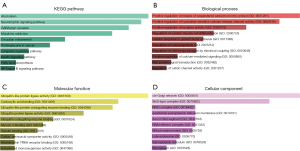
Enrichr database was used for KEGG pathway enrichment analysis and GO functional annotation of the totally 366 target genes. The top 10 enriched KEGG terms and GO items were shown in Figure 6, including Alcoholism, Neurotrophin signaling pathway and GABAergic synapse in KEGG; positive regulation of release of sequestered calcium ion into cytosol (GO:0051281), positive regulation of ryanodine-sensitive calcium-release channel activity (GO:0060316) and positive regulation of receptor activity (GO:2000273) in the BP category; ubiquitin-like protein ligase activity (GO:0061659), carboxylic acid binding (GO:0031406) and ubiquitin-like protein conjugating enzyme binding (GO:0044390) in the MF category; cis-Golgi network (GO:0005801), Sin3-type complex (GO:0070822) and RISC complex (GO:0016442) in the CC category.
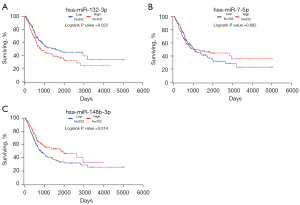
Kaplan-Meier survival curves were generated by the survival data from TCGA dataset, through OncoLnc. As shown in Figure 7, we found miR-132-3p and miR-148b-3p were related to the prognosis of BC patients, with P value =0.022 and 0.014, separately.
Discussion
BC is the 10th most common form of cancer worldwide (1), and the expenditure from diagnosis to death of per BC patient is the highest compared to other cancers (18). Early detection and timely intervention can improve patient survival and reduce the cost of treatment. Therefore, there is a need to find a novel and convenient diagnostic tool for BC. The miRNA screening based on serum is a convenient and widely applicable diagnostic method, especially noninvasive. A lot of studies have explored if the serum miRNAs serve as the diagnostic tool in BC (19,20). In our study, we paid more attention to BC detection and designed a three-stage study to evaluate the diagnostic performance of serum miRNAs in BC. In the beginning, we evaluated the expression level of 28 miRNAs in 5 BC pools and 3 HCs pools. Then, 10 dysregulated miRNAs chose as candidate miRNAs were confirmed in the training stage. After the three-stage study, five miRNAs in serum (miR-106a-5p, miR-145-5p, miR-132-3p, miR-7-5p and miR-148b-3p) were found significantly dysregulated between BC patient and HCs. After that, a three-miRNA panel, including miR-132-3p, miR-7-5p and miR-148b-3p, was set up and may be served as a potential serum marker for BC screening and diagnosis (AUC =0.922; 95% CI: 0.870 to 0.958; sensitivity =90.24%, specificity =81.71%; Figure 4).
In our study, we identified that miR-132-3p is a good serum biomarker for BC screening (AUC =0.781, 95% CI: 0.710 to 0.842; Figure 3F). There was previously reported that miR-132 is downregulated in BC tissues compared to adjacent normal tissues, and miR-132 might perform as tumor suppressor and, through TGFβ1/Smad2 signaling pathway, inhibit the invasion, migration and epithelial mesenchymal transition of BC cells (21). And, miR-132-3p may be sponged by circular RNAs DOCK1 (circDOCK1), and via modulation of circDOCK1/hsa-miR-132-3p/Sox5 pathway affect the progression of BC both in vitro and in vivo (22). All these studies showed that miR-132-3p might play a significant role in the development of BC.
Study has revealed that miR-7-5p was related to BC, and acted as a tumor suppressor by regulating the hedgehog pathway factor Gli3 in BC (23). And, miR-7 might be regulated by long non-coding RNA TINCR and facility BC progression via the TINCR/miR-7/mTOR signaling pathway (24). Du et al. identified a seven-miRNA panel including miR-7-5p that provided a relatively high diagnostic ability of BC with an AUC >0.9 in the study (25). Same as our study, miR-7-5p could serve as a biomarker for BC detection, and its diagnostic ability is 0.778 (95% CI: 0.707 to 0.839; Figure 3H).
In this study, miR-148b-3p could be served as a serum biomarker to discriminate BC patients from HCs alone (AUC =0.837, 95% CI: 0.771 to 0.890; Figure 3J), among the candidate miRNAs in the three-miRNA panel. Jiang et al. also focused on serum miRNA expression signatures for noninvasive diagnosis of BC and identified a six-miRNA panel including miR-148b-3p for the diagnosis of BC with an AUC of 0.899, through multivariate logistic regression model (26). In addition, exosome miR-148b-3p derived from cancer-associated fibroblasts might play the important role in the progression and chemosensitivity of BC and the study revealed that downregulated exosomal miR-148b-3p in cancer associated fibroblasts through downregulating the Wnt/β-catenin pathway and upregulating PTEN enhance chemosensitivity of BC cells (27).
BNC2, GAS7, and NTRK2 were considered as potential target genes of the three-miRNA panel, which were significantly dysregulated between BC patients and HCs, through GEPIA. Beothe et al. reported that they have detected novel tumor suppressor gene regions at chromosome 9p22.3p22 that harbor the BNC2 and PTPRD genes in multiplex urothelial carcinomas of the bladder by array comparative genomic hybridization (28). There have reported GAS7 deficiency promotes metastasis in MYCN-driven neuroblastoma (29). However, there are no studies revealing the role of GAS7 in BC, NTRK2 either. The role of GAS7 and NTRK2 in BC needs further exploration.
Although our results are promising, several limitations in this study should be addressed. First, as the sample size in this study is relatively small, it would be better to add external validation in large cohorts or different ethnic groups. Next, there are a lot of dysregulated miRNAs in the serum of patients with BC. However, only 28 miRNAs were included as the initial study, for the sake of avoiding the potential negligence of quantitative experiments and data processing. We will go on exploring the value of other miRNAs in serum in the diagnosis of BC and their biological effects.
Conclusions
In summary, the present study demonstrated five miRNAs that were significantly dysregulated in serum of BC patients, compared to HCs. Moreover, based on the diagnostic value of 3 candidate miRNAs (miR-132-3p, AUC =0.781; miR-7-5p, AUC =0.778; miR-148b-3p, AUC =0.837), the three-miRNA panel was set up to enhance the diagnostic efficacy of BC (AUC =0.922). Consequently, the three-miRNA panel, including miR-132-3p, miR-7-5p and miR-148b-3p, may be performed as a non-invasive, convenient and novel biomarker for the diagnosis of BC.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2611/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2611/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2611/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Peking University Shenzhen Hospital approved all study protocols (approval ID. 2017-007). All participants recruited in our study agreed to use their serum samples for research purposes and provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci 2019;110:408-19. [Crossref] [PubMed]
- Piatti P, Chew YC, Suwoto M, et al. Clinical evaluation of Bladder CARE, a new epigenetic test for bladder cancer detection in urine samples. Clin Epigenetics 2021;13:84. [Crossref] [PubMed]
- Almallah YZ, Rennie CD, Stone J, et al. Urinary tract infection and patient satisfaction after flexible cystoscopy and urodynamic evaluation. Urology 2000;56:37-9. [Crossref] [PubMed]
- Feber A, Dhami P, Dong L, et al. UroMark-a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics 2017;9:8. [Crossref] [PubMed]
- Uttley L, Whiteman BL, Woods HB, et al. Building the Evidence Base of Blood-Based Biomarkers for Early Detection of Cancer: A Rapid Systematic Mapping Review. EBioMedicine 2016;10:164-73. [Crossref] [PubMed]
- Ng K, Stenzl A, Sharma A, et al. Urinary biomarkers in bladder cancer: A review of the current landscape and future directions. Urol Oncol 2021;39:41-51. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One 2008;3:e3148. [Crossref] [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [Crossref] [PubMed]
- Fendler A, Stephan C, Yousef GM, et al. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol 2016;13:734-52. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67. [Crossref]
- Xie Z, Bailey A, Kuleshov MV, et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 2021;1:e90. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Botteman MF, Pashos CL, Redaelli A, et al. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics 2003;21:1315-30. [Crossref] [PubMed]
- Kutwin P, Borkowska EM, Bogucka P, et al. Expression profile of microRNAs (106b-3p, 130b-3, 145-3p, 199a-5p) in urine and serum samples from patients with the diagnosis of bladder cancer. Pol Merkur Lekarski 2021;49:103-7. [PubMed]
- Motawi TK, Rizk SM, Ibrahim TM, et al. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct 2016;34:142-8. [Crossref] [PubMed]
- Wei XC, Lv ZH. MicroRNA-132 inhibits migration, invasion and epithelial-mesenchymal transition via TGFβ1/Smad2 signaling pathway in human bladder cancer. Onco Targets Ther 2019;12:5937-45. [Crossref] [PubMed]
- Liu P, Li X, Guo X, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif 2019;52:e12614. [Crossref] [PubMed]
- Li J, Qiu M, An Y, et al. miR-7-5p acts as a tumor suppressor in bladder cancer by regulating the hedgehog pathway factor Gli3. Biochem Biophys Res Commun 2018;503:2101-7. [Crossref] [PubMed]
- Xu G, Yang H, Liu M, et al. lncRNA TINCR facilities bladder cancer progression via regulating miR 7 and mTOR. Mol Med Rep 2020;22:4243-53. [Crossref] [PubMed]
- Du L, Jiang X, Duan W, et al. Cell-free microRNA expression signatures in urine serve as novel noninvasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget 2017;8:40832-42. [Crossref] [PubMed]
- Jiang X, Du L, Wang L, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer 2015;136:854-62. [Crossref] [PubMed]
- Shan G, Zhou X, Gu J, et al. Downregulated exosomal microRNA-148b-3p in cancer associated fibroblasts enhance chemosensitivity of bladder cancer cells by downregulating the Wnt/β-catenin pathway and upregulating PTEN. Cell Oncol (Dordr) 2021;44:45-59. [Crossref] [PubMed]
- Beothe T, Zubakov D, Kovacs G. Homozygous losses detected by array comparative genomic hybridization in multiplex urothelial carcinomas of the bladder. Cancer Genet 2015;208:434-40. [Crossref] [PubMed]
- Dong Z, Yeo KS, Lopez G, et al. GAS7 Deficiency Promotes Metastasis in MYCN-Driven Neuroblastoma. Cancer Res 2021;81:2995-3007. [Crossref] [PubMed]


