
Copy number gains of chromosome 17 identified by dual in situ hybridization in non-small cell lung cancer tissue correlate with overexpression of c-Myc
Introduction
Non-small cell lung cancer (NSCLC) is one of the most common cancers globally and the leading cause of cancer-related death (1), urging a better understanding in molecular tumorigenesis and multidisciplinary treatment options. c-Myc, as a transcription and replication factor, regulates multiple genes that control cell growth in varied cancer types including NSCLC. Copy number gains of cytoband 17q25.3, along with chromosome 17, have been reported in patients having NSCLC, emphasizing its clinical significance as a potential molecular target for therapy in this cancer (2). In pre-clinical studies, the upregulation of c-Myc has been found to lead to accelerated tumor development associated with duplication of the syntenic human cytoband 17q25.3, emphasizing the important role of this region in tumorigenesis (3,4).
Fluorescence in situ hybridization (FISH) has been accepted as a standard assay in the assessment of gene status or copy number of a chromosome. However, the FISH assay has limitations, mainly related to the requirement of dark-field fluorescence microscopy and lack of morphologic information (5). An alternative brightfield method including dual in situ hybridization (DISH) comprising chromogenic and silver in situ hybridization (CISH and SISH, respectively), has been developed and demonstrated the advantage of being able to analyse tissue heterogeneity more comprehensively with acceptable concordance (5). This study aimed to explore and compare the correlations of chromosome 17 copy number identified by DISH assay and c-Myc expression in NSCLC with the paired-normal respiratory epithelium and to examine their role as potential molecular targets for NSCLC therapy. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2705/rc).
Methods
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Faculty of Medicine, Prince of Songkla University [protocol number REC 60-283-14-1]. Inform consent was waived.
Patient specimens and clinical information
This study included 66 cases of early-stage NSCLC (stages I–III) with paired-normal respiratory epithelial tissue. All patients had undergone radical surgery as their primary therapy. Tissue microarray slides were obtained from formalin-fixed, paraffin-embedded tissue sections, 3–4-µm in thickness, 3 mm in diameter (Figure 1). The copy number of the human epidermal growth factor receptor 2 (HER2) gene and chromosome 17 were evaluated by DISH. c-Myc protein expression was evaluated based upon immunohistochemistry (IHC) staining. Clinical information comprised the patient characteristics including demographic data, smoking status, NSCLC stage, histology, and survival outcome.
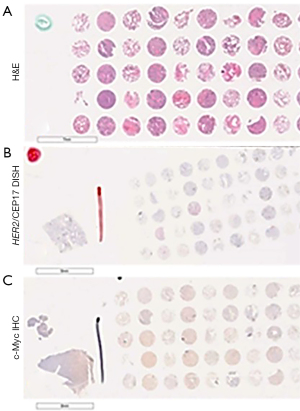
DISH
The copy number of chromosome 17 was determined by a HER2/centromeric enumeration probe of chromosome 17 (CEP17) bright-field DISH deoxyribonucleic acid (DNA) probe assay. DISH analysis was performed using a HER2 DISH DNA probe cocktail (Ventana Medical Systems, Tucson, AZ, USA). HER2 gene copy number was assessed using a SISH dinitrophenyl (DNP) detection kit, hapten labeling of the HER2 gene and red ISH digoxigenin (DIG) labeling of the CEP17. Briefly, unstained tissue microarray sections were baked overnight, deparaffinised in xylene, dehydrated in a series of ethanol washes, and air dried. The slides then were incubated in ribonuclease (RNase) for 60 minutes followed by a 10-minute cycle of protease digestion and denaturation. DNP- and DIG-labeled probes were cohybridized to their respective specific DNA sequences. The ultraView SISH DNP detection kit, followed by the Red ISH DIG detection kit were used to detect the DNP-labeled HER2 probe and the DIG-labeled chromosome 17 probe, respectively. The tissue microarray slides were then counterstained with hematoxylin II (Ventana Medical Systems). Image acquisition was performed using light microscopy. DISH counts were performed and the results were reported according to the established guideline. At least 100 non-overlapping intact interphase nuclei from each tissue sample were analyzed independently by 2 investigators (PS and Par T) to determine the average copy numbers and average ratios of HER2 gene and chromosome 17. The mean copy numbers of HER2 and chromosome 17 per cell were recorded, and the HER2:CEP17 ratio was calculated for each case. The following criteria adapted from the 2007 American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) criteria for dual-color ISH to classify a gain of HER2 gene or chromosome 17 was implemented.
Copy gains of HER2 gene: Gains were deemed in tissue specimens when the analysed nuclei presented an average ratio between the HER2 gene and the reference CEP17 probe (i.e., HER2:CEP17) ≥2.2.
Clonal gains of HER2 gene: Gains were defined when the analysed nuclei presented an average ratio between the HER2 gene and the reference CEP17 probe (i.e., HER2:CEP17) <2.2 (unmet criterion of the copy gains of HER2 gene), but ≥10% of the analysed nuclei demonstrated a HER2:CEP17 ratio ≥2.2.
Chromosome 17 copy gains or polysomy of chromosome 17: Gains of chromosome 17 were deemed when the analysed nuclei presented an average copy number ≥3 for both HER2 gene and CEP17, and the HER2:CEP17 ratio was <2.2.
IHC
Expression of c-Myc was determined by IHC. In brief, sections from the tissue microarray blocks were deparaffinized in xylene, rehydrated in graded alcohol, and washed in wash-buffer solution. Positive controls as well as negative reagent controls as a reference were included in each staining run as a reference. An epitope retrieval process using Dako Target Retrieval Solution (1:10; Agilent Technologies, Santa Clara, CA, USA) at 105 ℃ for 20 min was performed. The slides were then loaded onto an autostainer and the following incubations were applied sequentially: 10-min peroxide blocking agent (3%), 10-min Power Block, 10-min proteinase K, and a rabbit monoclonal anti-c-Myc antibody (1/50, EP121; Cell Marque, Rocklin, CA, USA). A buffer rinse was performed following each step. The slides were counterstained with hematoxylin, then rinsed gently in reagent quality water, dehydrated in graded alcohol, cleared in xylene, and cover-slipped.
Analyses of the IHC-stained slides were performed using the 10× magnification objective of a light microscope. c-Myc protein expression was categorized as positive or negative based on an IHC semi-quantitative score. In detail, the percentage of positive tumor cells per slide (0–100%) was multiplied by the dominant intensity pattern of staining (0= negative or trace; 1+ = weak; 2+ = moderate; 3+ = intense), giving an overall score ranging from 0–300. Specimens with IHC 2/3+ ≥75% of the cells of interest or an overall score ≥150 were considered positive for c-Myc.
Statistical analysis
Differences in c-Myc protein expression and chromosome 17 copy number gains between NSCLC patient subgroups were compared using univariate statistics, including the comparison of 2 means (t-test) and the comparison of 2 proportions (Chi-squared test). Multivariate statistics and Cox’s regression model to explore the relationship between c-Myc expression and the chromosome 17 copy number gains were also applied for the analysis. A P value of <0.05 was considered statistically significant. A Kaplan-Meier plotter and log-rank test were applied for survival analysis in each patient subgroup.
Results
The clinicopathological characteristics of the patients are shown in Table 1. The copy number gains of chromosome 17 were determined by HER2/CEP17 bright-field DISH DNA probe assay (Figure 2) and were found in 8 of 60 (13.3%) of the NSCLC specimens. No copy number gains of chromosome 17 were found in the paired-normal respiratory epithelium samples (Table 2). The mean HER2 (1.2±0.3) and CEP17 (1.4±0.3) copy numbers of the normal respiratory epithelium were significantly lower than those of the NSCLC tissue [1.8±1.0 and 2.0±0.8, respectively (P<0.001)]. However, the mean HER2/CEP17 ratios between the normal respiratory epithelium and the NSCLC tissue were not statistically significantly different (0.8±0.1 and 0.9±0.5, respectively (P=0.16; Table 3).
Table 1
| Clinicopathological characteristics | n (%) |
|---|---|
| Age (years), Mean ± SD | 61.8±10.9 |
| Age | |
| ≤65 years | 40 (60.6) |
| >65 years | 26 (39.4) |
| Gender | |
| Male | 37 (56.1) |
| Female | 29 (43.9) |
| Smoking status | |
| Smoker | 31 (47.0) |
| Never-smoker | 29 (43.9) |
| Unknown | 6 (9.1) |
| Tumor histology | |
| Adenocarcinoma | 46 (69.7) |
| Squamous carcinoma | 20 (30.3) |
| Clinical stage | |
| I | 43 (65.2) |
| II | 20 (30.3) |
| III | 3 (4.5) |
SD, standard deviation.
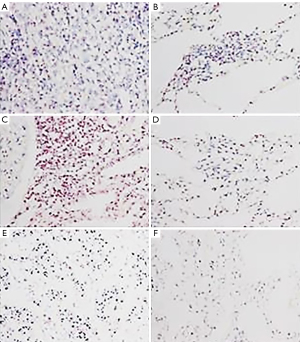
Table 2
| Normal respiratory epithelium, n (%) | NSCLC tissue, n (%) | P value* | Adenocarcinoma n=46, n (%) | Squamous carcinoma n=20, n (%) | P value* | |
|---|---|---|---|---|---|---|
| HER2 (DISH of HER2 and CEP17) | 0.001 | 0.013 | ||||
| Positive (HER2/CEP17 ratio ≥2.2) | 0 | 1 (1.7) | 1 (2.5) | 0 | ||
| Gains of HER2 and CEP17 (Copy number ≥3 and HER2/CEP17 ratio <2.2) | 0 | 8 (12.3) | 2 (5.0) | 6 (30.0) | ||
| Negative | 66 (100.0) | 51 (85.0) | 37 (92.5) | 14 (70.0) | ||
| Not available | 0 | 6 | 6 | 0 | ||
| cMyc expression (IHC 2/3+ ≥75% or score ≥150) | <0.001 | 0.005 | ||||
| Positive | 0 | 12 (18.2) | 4 (8.7) | 8 (40.0) | ||
| Negative | 66 (100.0) | 54 (81.8) | 42 (91.3) | 12 (60.0) | ||
| Not available | 0 | 0 | 0 | 0 | ||
*, Chi-square or Fisher exact test. HER2, human epidermal growth factor receptor 2; NSCLC, non-small cell lung cancer; DISH, dual-color in situ hybridization; CEP17, centromeric enumeration probe of chromosome 17; IHC, immunohistochemistry.
Table 3
| Normal respiratory epithelium, mean ± SD | NSCLC tissue, mean ± SD | P value* | |
|---|---|---|---|
| Mean HER2 copy number | 1.2±0.3 | 1.8±1.0 | <0.001 |
| Mean CEP17 copy number | 1.4±0.3 | 2.0±0.8 | <0.001 |
| Mean HER2/CEP17 ratio | 0.8±0.1 | 0.9±0.5 | 0.156 |
*, t-test. HER2, human epidermal growth factor receptor 2; SD, standard deviation; NSCLC, non-small cell lung cancer; CEP17, centromeric enumeration probe of chromosome 17.
Twelve of 66 (18.2%) NSCLC patients had overexpressed c-Myc protein detected by IHC. Five (41.7%) c-Myc positive patients also had HER2 gene amplification (1) or copy number gains of chromosome 17 (4), identified by gains of both HER2 and CEP17 (P=0.13; Table 4 and Figure 3).
Table 4
| c-Myc Negative, n (%) | c-Myc Positive, n (%) | P value* | |
|---|---|---|---|
| HER2 positive or gains of HER2 and CEP17 (polysomy) | 10 (18.5) | 5 (41.7) | 0.125 |
| HER2 negative | 44 (81.5) | 7 (58.3) | – |
*, Chi-square or Fisher exact test. HER2, human epidermal growth factor receptor 2; CEP17, centromeric enumeration probe of chromosome 17; CEP, centromeric enumeration probe.
The median HER2 and CEP17 copy numbers, as well as the median HER2/CEP17 ratios of the normal respiratory epithelium, were significantly lower than those of the NSCLC tissue, across all clinicopathological subgroups (Table 5). Multivariate analysis revealed that neither copy numbers of HER2 and chromosome 17 or any clinicopathological characteristics were independent predictors of overall survival in the NSCLC patients (Table 6).
Table 5
| n (%) | HER2 copy number | CEP17 copy number | HER2/CEP17 ratio | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal respiratory epithelium | NSCLC tissue | P value* | Normal respiratory epithelium | NSCLC tissue | P value* | Normal respiratory epithelium | NSCLC tissue | P value* | ||||
| Age | ||||||||||||
| ≤65 years | 40 (60.6) | 1.1 (1.0, 1.2) | 1.5 (1.2, 2.0) | <0.001 | 1.4 (1.2, 1.5) | 1.7 (1.4, 2.2) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.01 | ||
| >65 years | 26 (39.4) | 1.1 (1.0, 1.1) | 1.5 (1.3, 1.9) | <0.001 | 1.3 (1.2, 1.4) | 1.9 (1.6, 2.2) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.06 | ||
| Gender | ||||||||||||
| Male | 37 (56.1) | 1.1 (1.0, 1.2) | 1.5 (1.2, 2.0) | <0.001 | 1.3 (1.2, 1.4) | 1.8 (1.5, 2.3) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.006 | ||
| Female | 29 (43.9) | 1.1 (1.0, 1.2) | 1.6 (1.3, 1.9) | <0.001 | 1.3 (1.3, 1.4) | 1.8 (1.4, 2.0) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.12 | ||
| Smoking status | ||||||||||||
| Smoker | 31 (47.0) | 1.1 (1.0, 1.2) | 1.5 (1.2, 2.0) | <0.001 | 1.3 (1.2, 1.5) | 1.8 (1.5, 2.1) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.01 | ||
| Never-smoker | 29 (43.9) | 1.1 (1.1, 1.2) | 1.6 (1.2, 1.9) | <0.001 | 1.3 (1.2, 1.4) | 1.7 (1.4, 2.0) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.17 | ||
| Unknown | 6 (9.1) | 1 (1.1, 1.1) | 1.7 (1.3, 2.0) | 0.02 | 1.3 (1.3, 1.4) | 1.9 (1.5, 2.3) | 0.02 | 0.8 (0.8, 0.8) | 0.9 (0.8, 0.9) | 0.11 | ||
| Tumor histology | ||||||||||||
| Adenocarcinoma | 46 (69.7) | 1.1 (1.0, 1.2) | 1.4 (1.2, 1.6) | <0.001 | 1.3 (1.2, 1.4) | 1.6 (1.4, 1.9) | <0.001 | 0.9 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.09 | ||
| Squamous carcinoma | 20 (30.3) | 1.1 (1.0, 1.5) | 2.0 (1.5, 3.0) | <0.001 | 1.3 (1.3, 1.7) | 2.3 (1.9, 3.2) | <0.001 | 0.8 (0.8, 0.8) | 0.9 (0.8, 0.9) | 0.003 | ||
| Clinical stage | ||||||||||||
| I | 43 (65.2) | 1.1 (1.0, 1.2) | 1.5 (1.2, 1.9) | <0.001 | 1.3 (1.2, 1.4) | 1.7 (1.5, 2.1) | <0.001 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.07 | ||
| II | 20 (30.3) | 1.1 (1.1, 1.3) | 1.5 (1.3, 2.1) | 0.003 | 1.4 (1.3, 1.6) | 1.8 (1.5, 2.4) | 0.01 | 0.8 (0.8, 0.9) | 0.9 (0.8, 0.9) | 0.01 | ||
| III | 3 (4.5) | 1.0 (1.0, 1.0) | 1.5 (1.3, 1.8) | 0.35 | 1.3 (1.2, 1.4) | 1.9 (1.6, 2.1) | 0.4 | 0.8 (0.8, 0.8) | 0.8 (0.8, 0.9) | 0.27 | ||
Data are presented as median (IQR). *, t-test. HER2, human epidermal growth factor receptor 2; CEP17, centromeric enumeration probe of chromosome 17; IQR, interquartile range; NSCLC, non-small cell lung cancer.
Table 6
| Crude HR (95% CI) | Adjusted HR (95% CI) | P value (LR-test) | |
|---|---|---|---|
| Age: >65 vs. ≤65 years | 0.63 (0.28, 1.41) | 0.82 (0.32, 2.1) | 0.683 |
| Gender: female vs. male | 0.78 (0.36, 1.7) | 0.68 (0.16, 2.91) | 0.611 |
| Smoking status: Reference = Smoker | 0.769 | ||
| Never-smoker | 0.84 (0.38, 1.88) | 1.57 (0.34, 7.18) | |
| Unknown | 1.2 (0.34, 4.23) | 1.62 (0.36, 7.21) | |
| Tumor histology: squamous carcinoma vs. adenocarcinoma | 1.88 (0.87, 4.06) | 2.3 (0.8, 6.58) | 0.126 |
| Clinical Stage: Reference = Stage I | 0.112 | ||
| Stage II | 2.67 (1.23, 5.81) | 2.6 (1.06, 6.35) | |
| Stage III | 1.56 (0.2, 11.89) | 1.16 (0.13, 10.53) | |
| c-Myc tumor status: Positive vs. Negative | 1.46 (0.55, 3.89) | 1.21 (0.36, 4.14) | 0.76 |
| HER2 tumor status: Reference = Negative | 0.399 | ||
| Positive or polysomy | 1.06 (0.31, 3.57) | 0.56 (0.13, 2.37) | |
| Not available | 1.39 (0.47, 4.08) | 2.03 (0.6, 6.86) |
HER2, human epidermal growth factor receptor 2; HR, hazard ratio; CI, confidence interval; LR, likelihood ratio.
The prognostic significance of molecular features was evaluated by a Kaplan-Meier survival plotter and Cox proportional hazards model. NSCLC patients with HER2 gene amplification or copy number gain (polysomy) of chromosome 17 had a greater tendency to be associated with poorer overall survival than those with negative HER2 results, especially at 1 year [77.8 (95% CI: 36.5, 93.9) months vs. 96.1 (95% CI: 85.2, 99.0) months, respectively] and at 3 years [62.2 (95% CI: 21.3, 86.5) months vs. 71.1 (95% CI: 55.9, 81.9) months, respectively (P=0.954)] (Figure 4 and Table 7).
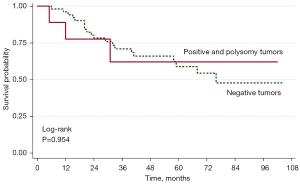
Table 7
| Variable | n | 1-year survival, % (95% CI) | 3-year survival, % (95% CI) | 5-year survival, % (95% CI) |
|---|---|---|---|---|
| HER2 (n=60) | ||||
| Negative | 51 | 96.1 (85.2, 99.0) | 71.1 (55.9, 81.9) | 58.9 (41.9, 72.4) |
| Positive and polysomy | 9 | 77.8 (36.5, 93.9) | 62.2 (21.3, 86.5) | 62.2 (21.3, 86.5) |
| c-Myc (n=66) | ||||
| Negative | 54 | 96.3 (86.0, 99.1) | 72.4 (57.7, 82.7) | 56.3 (40.0, 69.8) |
| Positive | 12 | 83.3 (48.2, 95.6) | 53.3 (19.9, 78.3) | 53.3 (19.9, 78.3) |
| HER2 and c-Myc (n=60) | ||||
| c-Myc and HER2 negative | 44 | 95.5 (83.0, 98.8) | 71.6 (55.3, 82.8) | 58.1 (39.9, 72.5) |
| c-Myc negative and HER2 positive | 4 | 100.0 (–, –) | 100.0 (–, –) | 100.0 (–, –) |
| c-Myc positive and HER2 negative | 7 | 100.0 (–, –) | 71.4 (25.8, 92.0) | 71.4 (25.8, 92.0) |
| c-Myc positive and HER2 positive | 5 | 60.0 (12.6, 88.2) | 40.0 (5.2, 75.3) | 40.0 (5.2, 75.3) |
HER2, human epidermal growth factor receptor 2; CI, confidence interval.
To determine whether the combination of c-Myc expression and HER2 copy number had a higher prognostic significance, 60 NSCLC patients whose tumors could be examined for both c-Myc and HER2 copy number were categorized based on the status of both variables. Patients with positive expression of c-Myc and HER2 amplification or gains of copy number of HER2 (co-positive results) tended to have a more unfavorable prognosis than the other subgroups (Figure 5 and Table 7).
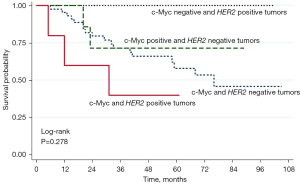
Discussion
NSCLC represents the leading cause of cancer-related deaths worldwide. Treatment outcome for NSCLC remains poor, indicating the need for better understanding of tumorigenesis and therapeutic development. c-Myc oncogene, the cellular homologue of the avian myelocytomatosis virus transforming sequence, encodes a nuclear transcription and replication factor. Alterations of c-Myc protein levels due to gene amplification, enhanced transcription, increased RNA or protein stability, chromosomal translocation, or viral insertion are involved in the initiation and progression of various cancer types (6). c-Myc stimulates the transcription of various target genes involved in cell growth and tumorigenesis and has been demonstrated to promote DNA replication via non-transcriptional mechanisms (7). Moreover, c-Myc has been reported to be associated with chemotherapy resistance and poor survival outcomes in various cancer types including NSCLC (8). In NSCLC, c-Myc has been recognized as an important oncogene in the initiation and progression of the tumor (9). Gains in copy numbers of c-Myc were found to be associated with lung adenocarcinoma in never-smokers (10). Increased activation of c-Myc has been observed in lung squamous cell carcinomas, compared to premalignant lesions in the same patients (11).
In this study, expression of c-Myc was significantly more demonstrable as positive IHC staining in tumorous tissue than normal respiratory tissue, confirming a role of c-Myc in NSCLC development. The expression of c-Myc was significantly less frequent in the adenocarcinoma subtype. In addition, NSCLC patients who had overexpressed c-Myc tended to have poorer overall survival than those who did not, highlighting the significance of this protein during stepwise lung carcinogenesis.
In a pre-clinical study, the upregulation of c-Myc lead to multiple changes in nuclear organization and selective chromosome repositioning including mouse chromosome 11 in PreB v-abl/myc cells, which were hypothesized to possibly promote neoplastic transformation including the mouse plasmacytoma (PCT). Accelerated tumor development has usually been associated with duplication of the mouse subcytoband 11E2 (syntenic to human chromosome 17, cytoband 17q25.3), supporting the importance of this chromosomal region in tumorigenesis (12). The 17q25 chromosomal region has been reported to be involved in a variety of human cancers including ovary (13,14), prostate (15), leukemia (16), neuroblastoma (17,18), esophagus (19-21), sarcoma (22,23), and breast (24). The frequent observations of allelic gain or loss in this chromosomal region in various types of cancers suggest that oncogenes or tumor suppressor genes, respectively, are located there. Frequent copy number gains of chromosome 17 have been proposed as a new phenomenon in NSCLC patients. Sunpaweravong et al. reported that gains of chromosome 17 were observed in approximately one third (38.9%) of NSCLC patients while a majority (61%) harbored clonal gains of cytoband 17q25.3. These patients also had a tendency towards poorer survival outcome (2).
HER2 has been well established as a driver and therapeutic target in several cancers including NSCLC (25). However, the term “HER2-positive NSCLC” represents a distinct HER2 aberration, not only at the molecular level but also in clinical entities and potential therapeutic agents. HER2 amplification has been reported in 10–20% of NSCLC patients while HER2 protein overexpression has been observed in 2.4-38% and HER2 mutations, such as in-frame insertions, have been detected in 2–4% (26-29). The Cancer Genome Atlas (TCGA) Research Network reports that tissue specimens from NSCLC patients harboring HER2 amplification have not been found to consistently associate with those carrying the HER2 mutation (27). Arcila et al. reported a group of 11 HER2-amplified lung cancer patients who had no co-existing HER2 mutations (30). However, the concurrence of HER2 amplifications and HER2 mutations have been reported in other studies, ranging from 3-50% of NSCLC patients (28,31,32).
Different HER2 detection methods and assessment criteria can impact HER2 results and interpretation. To detect HER2 amplification, in situ hybridization (ISH) remains the most used method. Both CISH and SISH have been accepted as a valid alternative to FISH and provide similar results in evaluating HER2 and CEP17 copy numbers (33). DISH and FISH are quantitatively-based techniques which have been used for the quantification of intracellular nucleic acids, however, FISH is more costly and time-consuming, requiring complex interpretation of fluorescency features. The major advantage of DISH images acquired from bright-field microscopy is higher degree in providing tissue morphology, especially for assessing tumor heterogeneity (5). We used the bright-field ISH assay to assess the copy numbers of HER2 and chromosome 17 in early-stage NSCLC patients. In this study, 13.3% of the available NSCLC specimens were found to have copy number gains of chromosome 17 while no copy number gains of chromosome 17 were observed in the paired-normal respiratory epithelium specimens. Additionally, the mean copy numbers of HER2 and CEP17 were also higher than those of the paired-normal respiratory epithelium specimens, emphasizing the important role of cancer-associated genes located on this chromosome which contribute to NSCLC tumorigenesis, including HER2/neu (17q12) in the EGFR family.
NSCLC has been characterized by extensive genomic instability at various levels, ranging from mutations in nucleotides to aneuploidy resulting from gains or losses of DNA copy numbers or entire chromosomes (8). Similar to our study, polysomy of chromosome 17 has been previously reported as a frequent phenomenon in NSCLC (25,31). However, apart from this study, the prognostic significance of polysomy of chromosome 17 has not been well established before. We, therefore, encourage further investigations to evaluate and confirm the potential prognostic and/or predictive value of chromosome 17 polysomy in NSCLC development and treatment outcomes.
We found that 41.7% of c-Myc positive NSCLC patients also had HER2 gene amplification or copy number gains of the entire chromosome 17, identified by gains of both HER2 and CEP17. Additionally, we found that NSCLC patients who harbored the HER2 gene amplification or had copy number gains of chromosome 17 and positive expression of c-Myc tended to be associated with poor overall survival. The results from this study suggest that copy number gains of chromosome 17 and c-Myc overexpression could be promising prognostic biomarkers for NSCLC. However, this study is a preliminary report of a limited number of tumor samples, therefore further investigations are required to confirm our findings.
Conclusions
The HER2 DISH DNA probe provides a reliable way to detect the chromosome 17 copy number in NSCLC patients and could be considered as an alternative assay to FISH. We found that copy number gains of chromosome 17 correlated with overexpression of c-Myc, emphasizing an important genetic aberration on this chromosome in NSCLC tumorigenesis. Both biomarkers may be used as prognostic factors to identify NSCLC patients with poorer survival outcomes and are potential targets for therapeutic development.
Acknowledgments
Funding: This work was funded by the Faculty of Medicine, Prince of Songkla University.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2705/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2705/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2705/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Faculty of Medicine, Prince of Songkla University (protocol number REC 60-283-14-1). Inform consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Sunpaweravong P, Thu KL, Lam WL, et al. Assessment of the clinical relevance of 17q25.3 copy number and three-dimensional telomere organization in non-small lung cancer patients. J Cancer Res Clin Oncol 2016;142:749-56. [Crossref] [PubMed]
- Mai S, Garini Y. Oncogenic remodeling of the three-dimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell Cycle 2005;4:1327-31. [Crossref] [PubMed]
- Mai S. Initiation of telomere-mediated chromosomal rearrangements in cancer. J Cell Biochem 2010;109:1095-102. [Crossref] [PubMed]
- Gao FF, Dabbs DJ, Cooper KL, et al. Bright-field HER2 dual in situ hybridization (DISH) assay vs fluorescence in situ hybridization (FISH) focused study of immunohistochemical 2+ cases. Am J Clin Pathol 2014;141:102-10. [Crossref] [PubMed]
- Kuzyk A, Mai S. c-MYC-induced genomic instability. Cold Spring Harb Perspect Med 2014;4:a014373. [Crossref] [PubMed]
- Dominguez-Sola D, Ying CY, Grandori C, et al. Non-transcriptional control of DNA replication by c-Myc. Nature 2007;448:445-51. [Crossref] [PubMed]
- Varella-Garcia M. Chromosomal and genomic changes in lung cancer. Cell Adh Migr 2010;4:100-6. [Crossref] [PubMed]
- Chen S, Xu Y, Chen Y, et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS One 2012;7:e36326. [Crossref] [PubMed]
- El-Telbany A, Ma PC. Cancer genes in lung cancer: racial disparities: are there any? Genes Cancer 2012;3:467-80. [Crossref] [PubMed]
- Ooi AT, Gower AC, Zhang KX, et al. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev Res (Phila) 2014;7:487-95. [Crossref] [PubMed]
- Wiener F, Schmälter AK, Mowat MR, et al. Duplication of Subcytoband 11E2 of Chromosome 11 Is Regularly Associated with Accelerated Tumor Development in v-abl/myc-Induced Mouse Plasmacytomas. Genes Cancer 2010;1:847-58. [Crossref] [PubMed]
- Presneau N, Dewar K, Forgetta V, et al. Loss of heterozygosity and transcriptome analyses of a 1.2 Mb candidate ovarian cancer tumor suppressor locus region at 17q25.1-q25.2. Mol Carcinog 2005;43:141-54. [Crossref] [PubMed]
- Wojnarowicz PM, Provencher DM, Mes-Masson AM, et al. Chromosome 17q25 genes, RHBDF2 and CYGB, in ovarian cancer. Int J Oncol 2012;40:1865-80. [PubMed]
- Bermudo R, Abia D, Benitez D, et al. Discovery of genomic alterations through coregulation analysis of closely linked genes: a frequent gain in 17q25.3 in prostate cancer. Ann N Y Acad Sci 2010;1210:17-24. [Crossref] [PubMed]
- Gulten T, Yakut T, Karkucak M, et al. AML1 amplification and 17q25 deletion in a case of childhood acute lymphoblastic leukemia. J Clin Lab Anal 2009;23:368-71. [Crossref] [PubMed]
- Islam A, Kageyama H, Hashizume K, et al. Role of survivin, whose gene is mapped to 17q25, in human neuroblastoma and identification of a novel dominant-negative isoform, survivin-beta/2B. Med Pediatr Oncol 2000;35:550-3. [Crossref] [PubMed]
- Yu M, Ohira M, Li Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol 2009;34:931-8. [PubMed]
- Risk JM, Evans KE, Jones J, et al. Characterization of a 500 kb region on 17q25 and the exclusion of candidate genes as the familial Tylosis Oesophageal Cancer (TOC) locus. Oncogene 2002;21:6395-402. [Crossref] [PubMed]
- Langan JE, Cole CG, Huckle EJ, et al. Novel microsatellite markers and single nucleotide polymorphisms refine the tylosis with oesophageal cancer (TOC) minimal region on 17q25 to 42.5 kb: sequencing does not identify the causative gene. Hum Genet 2004;114:534-40. [Crossref] [PubMed]
- McRonald FE, Liloglou T, Xinarianos G, et al. Down-regulation of the cytoglobin gene, located on 17q25, in tylosis with oesophageal cancer (TOC): evidence for trans-allele repression. Hum Mol Genet 2006;15:1271-7. [Crossref] [PubMed]
- Ladanyi M, Lui MY, Antonescu CR, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 2001;20:48-57. [Crossref] [PubMed]
- Uppal S, Aviv H, Patterson F, et al. Alveolar soft part sarcoma-reciprocal translocation between chromosome 17q25 and Xp11. Report of a case with metastases at presentation and review of the literature. Acta Orthop Belg 2003;69:182-7. [PubMed]
- Morris CM, Haataja L, McDonald M, et al. The small GTPase RAC3 gene is located within chromosome band 17q25.3 outside and telomeric of a region commonly deleted in breast and ovarian tumours. Cytogenet Cell Genet 2000;89:18-23. [Crossref] [PubMed]
- Li BT, Ross DS, Aisner DL, et al. HER2 Amplification and HER2 Mutation Are Distinct Molecular Targets in Lung Cancers. J Thorac Oncol 2016;11:414-9. [Crossref] [PubMed]
- Heinmöller P, Gross C, Beyser K, et al. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res 2003;9:5238-43. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Suzuki M, Shiraishi K, Yoshida A, et al. HER2 gene mutations in non-small cell lung carcinomas: concurrence with HER2 gene amplification and HER2 protein expression and phosphorylation. Lung Cancer 2015;87:14-22. [Crossref] [PubMed]
- Kim EK, Kim KA, Lee CY, et al. The frequency and clinical impact of HER2 alterations in lung adenocarcinoma. PLoS One 2017;12:e0171280. [Crossref] [PubMed]
- Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910-8. [Crossref] [PubMed]
- Li C, Sun Y, Fang R, et al. Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol 2012;7:85-9. [Crossref] [PubMed]
- Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997-2003. [Crossref] [PubMed]
- Francis GD, Jones MA, Beadle GF, et al. Bright-field in situ hybridization for HER2 gene amplification in breast cancer using tissue microarrays: correlation between chromogenic (CISH) and automated silver-enhanced (SISH) methods with patient outcome. Diagn Mol Pathol 2009;18:88-95. [Crossref] [PubMed]


