
Comparative study on the mutation spectrum of tissue DNA and blood ctDNA in patients with non-small cell lung cancer
Introduction
Lung cancer is a malignant tumor with the highest mortality and second highest morbidity worldwide. In 2020, there were 2.2 million new cases of lung cancer worldwide and 1.8 million deaths (1). Non-small cell lung cancer (NSCLC) treatment methods include surgical resection, comprehensive adjuvant treatment, neoadjuvant radiotherapy, and chemotherapy. Surgery and puncture tissue samples to detect gene mutations are still the gold standard for diagnosing whether NSCLC patients are suitable for targeted therapy (2). The sooner the patients receive treatment, the better the treatment effect. However, most NSCLC patients are at the middle or advanced stage when the diagnosis is confirmed, losing the opportunity for surgical treatment. Traditional treatment uses platinum-containing dual-agent chemotherapy as the first-line treatment. The National Comprehensive Cancer Network (NCCN) treatment guidelines recommend that patients with advanced NSCLC take comprehensive treatment based on systemic therapy (3). Although chemotherapy has made significant progress, the overall therapeutic effect of NSCLC is still not satisfactory, and its 5-year survival rate remains low.
There are two types of mutations in the tumor genome: “passenger mutations”, which have little effect on the development of tumors, gradually disappear passively as the disease progresses. “Driver mutation” is another type of mutation that is crucial to oncogenesis and tumor progression. Cancer cells carrying driver mutations aberrantly activate/inhibit key signaling pathways and have selective advantages in survival or proliferation (4). Since tumors rely on driver mutations to maintain the malignant phenotype, it has also become a hot spot in current tumor treatment research. Recent studies have shown that NSCLC has a significant connection with gene mutations. The commonly NSCLC driver genes include epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), Kirsten rat sarcoma viral oncogene (KRAS), phosphatidylinositol 3-kinase catalytic subunit α (PIK3CA), and mesenchymal to epithelial transition factor (MET), which mutated may directly “drive” the oncogenesis and tumor progression of NSCLC (5-7). Currently, EGFR-TKIs are widely used in the treatment regimen of patients with EGFR-mutant NSCLC while benefiting patients (8).
The diagnosis of NSCLC mainly relies on pathological biopsy (9). Although this traumatic examination method can directly observe pathological cell changes, it has poor reproducibility and is not suitable for post-treatment monitoring. Many patients lose the opportunity for targeted therapy due to the inability to obtain tissue specimens or too few tissue specimens are obtained for molecular type testing. At the same time, due to the existence of tumor heterogeneity, mutations of different tumor sites and even different tumor cells at the same tumor site may be different (10), and the genetic testing of a single tissue biopsy cannot accurately reflect the overall characteristics of the tumor (11). With the evolution of tumors caused by treatment selection pressure and the emergence of drug-resistant genes, relying only on pre-treatment specimens to guide subsequent treatment may lead to treatment bias (12).
ctDNA is tumor-derived fragmented DNA in the blood. ctDNA comes directly from the tumor or from circulating tumor cells and usually carries the gene mutation information of the tumor tissue. In recent years, ctDNA-based liquid biopsy has provided new opportunities for molecular diagnosis and monitoring of tumors. ctDNA gene mutation detection can be used for early cancer screening and real-time monitoring of cancer development, metastasis, and prognosis (13). Circulating tumor DNA (ctDNA) is a tumor-specific gene carried by apoptotic tumor cells and is released into the blood, which can fully reflect the molecular composition of tumor tissue. Lebofsky et al. confirmed that 97% of mutation genes in metastasis biopsies could be found in ctDNA of tumor-matched samples (14). Guo et al. revealed that NGS detection results of tumor tissue samples and ctDNA samples are not entirely consistent. The concordance was 54.6% in patients with early-stage NSCLC and 80% in patients with advanced NSCLC, so indifferent patients, the intrinsic biological Mechanisms may affect the consistency of tumor biopsies and ctDNA genomic profiles (15). Therefore, this study intends to use NGS panel testing to detect tissue samples and ctDNA samples to analyze the mutation type and mutation frequency of differential genes and differences in the enrichment of biological functions of mutant genes. NSCLC patients with EGFR sensitive mutations respond well to EGFR-tyrosine kinase inhibitor (TKI) treatment, but most patients develop resistance 9–13 months after receiving treatment (16). With the emergence of new targeted drugs and the in-depth study of drug resistance mechanisms, a more comprehensive understanding of the composition of patients’ oncogenes is needed to guide treatment and evaluation.
This study analyzed the detection results of gene mutations in tissue samples and blood samples of patients with advanced NSCLC, explored the relationship between ctDNA detection and prognostic judgment, and performed curative effect evaluation and recurrence monitoring of NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-970/rc).
Methods
Patient enrollment
The cohort included 28 NSCLC patients from Guangzhou First People’s Hospital. Among them, 11 patients underwent complete tumor staging in accordance with the seventh edition of the NSCLC tumor, lymph node, and metastasis (TNM) standards. Tissue samples were obtained through prostate puncture or radical prostate cancer surgery, then tissues were paraffin-embedded, sliced, and submitted for examination. Blood samples were collected from the patient’s vein and stored in an EDTA anticoagulant tube for cryopreservation, then submitted for inspection. All samples submitted for inspection met the quality control requirements of gene sequencing and were sequenced for mutation analysis. This study has been approved by the relevant supervision and independent ethics committee of Guangzhou First People’s Hospital (No. K-2020-051-01). Every patient provided written informed consent for use of their samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Next-generation sequencing (NGS) and annotation
DNA was isolated from the FFPE samples and blood samples using the DNeasy Blood and Tissue Kit (69504, QIAGEN, Venlo, Netherlands). Multi-gene panel targeted NGS including 556 genes was performed on Ion Torrent (Tongshu BioTech, Shanghai, China). The targeted libraries were constructed using NGS Fast DNA Library Prep Set (Thermo Fisher, Waltham, MA, USA). BWA (Burrows-Wheeler-Alignment) software was used to compare the sequencing results. GATK (The Genome Analysis Toolkit) was used to correct the comparison quality. Mutect2 and VarDict software were used to detect somatic mutation sites from the comparison results, and the intersection of mutation sites detected by the software was used as the candidate somatic mutation sites. For the detected candidate somatic mutation sites, ANNOVAR software was used to annotate the population frequency in the ExAC (Exome Aggregation Consortium) and ESP (NHLBI Exome Sequencing Project) databases and the functional information of mutation function and mutation type. In order to further improve the credibility of mutations, we removed mutants with low support for reads according to the sequencing depth of the loci. In addition, mutation sites with a population frequency greater than 0.01 were also removed, and the remaining mutations were used as mutation sites with high confidence for subsequent analysis.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
Oncoplot mutations were plotted for each sample and different groupings using the MAfTools R package. Mutations in cancer-related driver genes were also analyzed. The clusterProfiler R package was also used for GO and KEGG functional enrichment analysis of mutated genes in each grouping sample. In order to analyze the overlap between samples of different groups, the Venn diagram R package was also used to draw Venn diagrams between each group of mutations.
Statistical analysis
GraphPad Prism 8.0 software was used for statistical analysis. The t-test was used for continuous variables, while the chi-square test or Fisher’s exact test was used for categorical variables. Two-sided tests were used, and P<0.05 was considered statistically significant.
Results
Patient characteristics
From May 2018 to Dec 2020, we recruited 28 patients diagnosed with advanced or metastatic NSCLC and collected 39 samples (24 tumor tissues and 15 blood samples) from the patients. In addition, paired tissue samples and blood samples were tested in 11 patients. The median age of the 11 patients was 64 years (55–77 years). Six cases were females, and 5 cases were males. Four patients had a history of smoking, while 7 did not smoke. Nine patients were diagnosed with adenocarcinoma and 2 patients with squamous cell carcinoma. Three patients accepted EGFR-TKI treatment, and 2 patients accepted chemotherapy. There was a lack of treatment information for the other patients. Table 1 summarizes the clinical characteristics of the cohort.
Table 1
| No. | Gender | Age (years) | Smoking | Clinical stage | Treatment | Therapeutic evaluation | Metastasis | Pathology |
|---|---|---|---|---|---|---|---|---|
| Pt.1 | Male | 64 | Yes | IVB | – | – | Lymph nodes, bone, kidney, pleura | Squamous carcinoma |
| Pt.2 | Female | 60 | None | IVA | – | – | Pleura | Adenocarcinoma |
| Pt.3 | Male | 58 | Yes | IVB | – | – | Lymph nodes, pleura, pelvic cavity | Adenocarcinoma |
| Pt.4 | Female | 55 | None | IVB | – | – | Lymph nodes, bone | Adenocarcinoma |
| Pt.5 | Female | 77 | None | IVB | EGFR-TKI | PR | Lymph nodes, bone, liver | Adenocarcinoma |
| Pt.6 | Male | 75 | None | IVB | EGFR-TKI | PR | Lymph nodes, bone, kidney, brain | Adenocarcinoma |
| Pt.7 | Female | 60 | None | IVB | Chemotherapy | SD | Lymph nodes, bone, kidney, liver | Adenocarcinoma |
| Pt.8 | Female | 72 | None | IVA | EGFR-TKI | PR | Lymph nodes, pleura | Adenocarcinoma |
| Pt.9 | Female | 67 | None | IVA | Chemotherapy, Tislelizumab | PR | Lymph nodes, pleura | Squamous carcinoma |
| Pt.10 | Male | 69 | Yes | IVA | – | – | Lymph nodes | Adenocarcinoma |
| Pt.11 | Male | 60 | Yes | IVA | – | – | Nodes, pleura | Adenocarcinoma |
EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; PR, partial response; SD, stable disease.
Comparison of the mutational profiles of tissue DNA and ctDNA
We analyzed a total of 39 mutation profiles of tumor tissues and ctDNA samples through NGS detection. Overall, 145 genes were found to have mutations in tumor tissues and ctDNA samples, including 28 deletions (DELs), 7 insertions (INSs), and 270 single nucleotide variants (SNVs). The top 3 mutant genes were TP53 (59.0%, 23/39), EGFR (33.3%, 13/39), and LRP1B (23.1%, 9/39) (Figure 1A). A total of 114 genes were found to have mutations in tissue samples, including 9 Frame_Shift_Del, 2 Frame_Shift_Ins, 7 In_Frame_Del, 1 In_Frame_Ins, 157 missense mutations, 16 nonsense mutations, and 1 Splice_Site (Table S1). The common mutated genes were TP53 (58.3%, 14/24), EGFR (33.3%, 8/24), LRP1B (25.0%, 6/24), KRAS (20.8%, 5/24), and ARID1A (16.7%, 4/24) (Figure 1B). A total of 78 genes were found to have mutations in ctDNA samples, including 4 Frame_Shift_Del, 2 Frame_Shift_Ins, 5 In_Frame_Del, 1 In_Frame_Ins, 85 missense mutations, 13 nonsense mutations, and 2 Splice_Site (Table S2). The top 3 mutant genes were TP53 (60%, 5/15), EGFR (33.3%, 5/15), LRP1B (20.0%, 3/15), ERBB4 (3/15), and ARID1A (13.3%, 2/15) (Figure 1C).
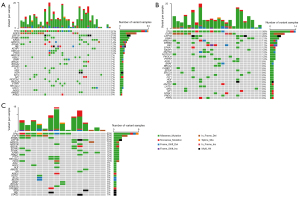
The relationship between tissue DNA and ctDNA in NSCLC patients
Among the 28 patients, 11 patients underwent both tissue sample DNA testing and blood ctDNA testing. As shown in Table 2, there were 100 mutated genes detected in tissue and ctDNA samples, of which 39% were co-mutated genes (39%, 39/100). Among them, 72 mutated genes were detected in tissue samples, and 33 (45.8%, 33/72) mutated genes were ctDNA-specific. The ctDNA samples detected mutations in 67 genes, and 28 (41.8%, 28/67) mutated genes were ctDNA-specific. In addition, 39 genes were co-mutated in both samples, tissue samples accounted for 54.2% (39/72), and ctDNA samples accounted for 58.2% (39/67). The average co-mutation frequency of paired tissue DNA with ctDNA was 40.1% (0–83.3%), and the median value was 37.5%. The average accuracy of paired tissue DNA with ctDNA detection was 65.26% (0–100%), and the median value was 90.91% (Table 3).
Table 2
| Samples | Mutation genes | Total (n=100) |
|---|---|---|
| Tissue | ERCC4, FLT4, ATRX, HIST1H3C, MAP3K13, FOXL2, LATS1, MCL1, KMT2C, PTPRT, EP300, CARD11, SDHA, NRAS, PTPRS, WWTR1, ERCC3, ROS1, SHOC2, MSH3, KMT2D, SDHAF2, MET, PREX2, ATR, CIC, PIK3C2G, PIK3CA, FGF10, SGK1, KLF4, SNCAIP, FAT1 | 33 |
| ctDNA | ARID2, CYSLTR2, HOXB13, STAG2, FOXP1, LTK, SUZ12, KMT2B, PTPN11, FANCL, TNFRSF14, ELF3, PIK3CG, STAT5A, SPEN, RECQL4, PRKD1, SRC, SF3B1, TSHR, CDKN1A, NOTCH3, ABL1, ALK, RASA1, AR, ERBB2, KDM6A | 28 |
| Tissue and ctDNA | EPHA5, BRCA1, PRKCI, ERBB4, E2F3, KRAS, EPHA7, BRAF, IRS2, TCF3, LRP1B, TERT, KDR, HGF, TP53, ZFHX3, PTEN, ASXL1, RET, ARID1A, BCL6, STK11, NF1, RAF1, CTNNB1, AMER1, ETV6, EGFR, FGF12, MYC, MGA, AGO2, BCOR, KEAP1, NCOR1, BRCA2, POLD1, PARP1, ARID1B | 39 |
ctDNA, circulating tumor DNA.
Table 3
| Patients | Tissue specific mutation | Common mutation | ctDNA specific mutation | Co-mutation frequency | Accuracy |
|---|---|---|---|---|---|
| Pt.1 | 3 | 10 | 1 | 71.43% | 90.91% |
| Pt.2 | 2 | 7 | 0 | 77.78% | 100.00% |
| Pt.3 | 1 | 5 | 0 | 83.33% | 100.00% |
| Pt.4 | 0 | 2 | 4 | 33.33% | 33.33% |
| Pt.5 | 3 | 6 | 3 | 50.00% | 66.67% |
| Pt.6 | 1 | 0 | 1 | 0.00% | 0.00% |
| Pt.7 | 3 | 2 | 0 | 40.00% | 100.00% |
| Pt.8 | 6 | 1 | 17 | 4.17% | 5.56% |
| Pt.9 | 10 | 6 | 0 | 37.50% | 100.00% |
| Pt.10 | 16 | 3 | 11 | 10.00% | 21.43% |
| Pt.11 | 6 | 3 | 0 | 33.33% | 100.00% |
ctDNA, circulating tumor DNA.
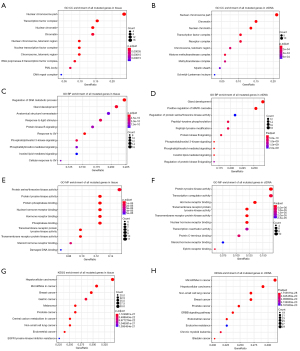
Next, we performed GO and KEGG analysis to assess the mutated genes in tissue and ctDNA samples. This study found that for the co-enrichment of tissue and ctDNA samples, the GO terms of cellular component (CC) were nuclear chromosome part, chromatin, nuclear chromatin, transcription factor complex, and chromosome/telomeric region (Figure 2A,2B). The GO terms of biological process (BP) were gland development, phosphatidylinositol 3-kinase signaling, inositol lipid-mediated signaling, regulation of protein kinase B signaling, phosphatidylinositol-mediated signaling, and protein kinase B signaling (Figure 2C,2D). The GO terms of molecular function (MF) were protein tyrosine kinase activity, hormone receptor binding, transmembrane receptor protein tyrosine kinase activity, transmembrane receptor protein kinase activity, nuclear hormone receptor binding, and steroid hormone receptor binding (Figure 2E,2F). In addition, the KEGG pathway enrichment analysis showed that the mutated gene functions in tissue and ctDNA were mainly enriched in hepatocellular carcinoma, NSCLC, breast cancer, prostate cancer, endometrial cancer, and tumor-associated miRNA (Figure 2G,2H).
As showing Table 1, the follow-up treatment information was available for 5 of the 11 patients who had both tissue and ctDNA samples tested, including 3 who received EGFR-TKI therapy (all patients evaluated as partial response) and 2 who received chemotherapy (one evaluated as stable and the other as partial response). EGFR mutations were found in tissue samples and ctDNA samples from patients treated with EGFR-TKIs. In contrast, tissue samples and ctDNA samples from chemotherapy patients did not have EGFR mutations.
Discussion
We performed NGS testing on 28 patients with lung cancer, including 24 tissue samples and 15 blood samples. In addition, 11 of the 28 patients received tissue sample testing and ctDNA sample testing. The analysis of the sequencing results revealed some crucial questions. For example, ctDNA testing found that patients with lung cancer carry tumor-related gene mutations, such as TP53, KRAS, and EGFR mutations. This detection has important clinical significance, suggesting that ctDNA mutation detection can be used as a non-invasive early detection method and can be applied to screening high-risk populations. In the process of cancer screening and early diagnosis, the combination of the quantitative level of ctDNA in circulation and the identification of ctDNA gene mutations can provide valuable information for cancer diagnosis. Existing studies indicate that gene mutations are tumor-specific and detection of mutations in local tumor tissues is often insufficient (17,18). Assessing these changes in ctDNA can provide greater diagnostic accuracy than standard protein biomarkers such as carcinoembryonic antigen (CEA) (19,20).
ctDNA from tumors can represent the entire genome pattern of tumors to a certain extent and can be used as a liquid biopsy to analyze tumor-specific genetic and epigenetic changes, including oncogene or tumor suppressor gene mutations, methylation, and gene amplification, among others. As an effective and reliable biomarker in the past few years, ctDNA has played a vital role in classifying specific patients, guiding early clinical diagnosis and treatment, and for prognostic evaluation. In addition, using ctDNA biomarkers to detect epigenetic abnormalities in specific types of cancers early, such as hypomethylation of promoters in oncogenes or tumor suppressor genes, can provide critical information for tumor biology research and clinical treatment. Several studies have shown that TP53, ITH, HCK, and TNNB1 are common in peripheral blood ctDNA in patients with liver cancer (21-24). Specific mutations suggest that peripheral blood ctDNA can be detected for the early diagnosis of cancerous hepatocytes. Gormally et al. reported that plasma ctDNA could detect KRAS gene mutations in healthy subjects as early as 2 years in advance and could detect the mutation of TP53 in healthy patients 20.8 months before cancer diagnosis (25). Although the plasma of advanced patients with early tumors or tumor micrometastasis contains only a relatively low abundance of ctDNA fragments, quantitative and qualitative analysis of trace amounts of DNA has become possible with the continuous advancement of DNA detection technology (26).
The prognosis of cancer patients is usually evaluated by combining clinical observation, tumor progression stage, histopathological changes, and biomolecular features. When tissue sections are challenging to obtain, such as non-solid tumors, or it is difficult to use preserved tissue sections for genetic analysis, the importance of using patient blood to detect ctDNA is revealed. In addition, due to tumor heterogeneity, ctDNA detection can reflect the mutation information of patients more comprehensively, while tissue DNA detection often reflects the mutation information of a single nodule. ctDNA detection can dynamically detect the prognosis of patients during treatment and support timely adjustment of patient treatment regimens. Due to the advantage of ctDNA detection. Due to the potential benefits of ctDNA detection, many studies reveal the feasibility of ctDNA detection as a tumor liquid biopsy. The study of Bettegowda et al. showed that ctDNA could be detected in more than 75% of NSCLC, breast cancer, ovarian cancer, pancreatic cancer, colorectal cancer, liver cancer, and head and neck cancer (27). Studies have shown that ctDNA can reveal a series of molecular biological characteristics related to tumors, such as RAS gene and TP53 gene mutations, APC gene hypermethylation, loss of allelic heterozygosity (LOH), and mtDNA changes (28-30). About one-third of NSCLC patients have KRAS gene mutations. Gautschi et al. studied 180 cases of lung cancer patients with ctDNA and found that KRAS mutations are associated with poor prognosis (31). Parkinson et al. analyzed the mutations of TP53 gene ctDNA in 40 patients with malignant, recurrent, high-grade serous ovarian cancer (32). They showed that after 1 course of chemotherapy, patients whose TP53 mutation allele scores decreased >60% had a more prolonged disease-relapse survival time. Morikawa et al. tested plasma PIK3CA and KRAS gene ctDNA in patients with ovarian clear cell carcinoma and showed that patients with high PIK3CA-H1047R or KRAS-G12D ctDNA had a shorter survival time (33). A series of studies have shown that the plasma ctDNA of patients with pancreatic cancer contains mutations in the KRAS gene. Mutations in the KRAS gene can monitor the development of pancreatic cancer (34,35). Dabritz et al. detected 56 cases of pancreatic ductal adenocarcinoma and 13 cases of pancreatitis with plasma ctDNA mutations in the KRAS gene (36). Among them, found KRAS mutation in 20 patients with pancreatic duct adenocarcinoma, but no KRAS mutation in patients with pancreatitis. In a study of colorectal cancer, Trojan et al. compared KRAS gene mutation and CDKN2A promoter hypermethylation in the blood ctDNA of 37 patients with colorectal cancer. They found that the KRAS gene had mutations or CDKN2A promoters. Patients with hypermethylation had a 2-year survival rate of 48%, while those without changes had a 2-year survival rate of 100% (37).
However, one limitation of ctDNA detection is that the low abundance of ctDNA fragments in blood makes its detection sensitivity challenging. Another, the results of ctDNA assays may be interfered with by clonal hematopoietic and germline mutations. Despite this, ctDNA detection has a good application prospect in the clinical diagnosis and treatment of tumors. For liquid biopsy to detect whether patients contain ctDNA and gene mutations is conducive to an early screening of patients and timely early intervention. For localized cancers, ctDNA testing can help doctors improve patient treatment plans. At the same time, during the treatment process, ctDNA detection can dynamically monitor tumor burden and curative effect. After treatment, blood ctDNA content and gene mutation can still be detected in real-time to monitor cancer recurrence and metastasis.
Conclusions
This study sequenced tissue and ctDNA samples from 28 patients with advanced NSCLC by targeted NGS. ctDNA detection was consistent with tumor tissue detection to a certain extent. In addition, patients with EGFR mutations detected in both tissue samples and ctDNA samples achieved partial remission after EGFR-TKI treatment. ctDNA detection in patients as a method of disease diagnosis and evaluation of tumor molecular status is advantageous in clinical diagnosis and treatment.
Acknowledgments
The authors are grateful to Shanghai Tongshu Biotechnology Co., Ltd. for technical support.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-970/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-970/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-970/coif). All authors report the technical support from Shanghai Tongshu Biotechnology Co., Ltd. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study has been approved by the relevant supervision and independent ethics committee of Guangzhou First People’s Hospital (No. K-2020-051-01). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all study participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 2.2021. J Natl Compr Canc Netw 2021;19:254-66. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Osmani L, Askin F, Gabrielson E, et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin Cancer Biol 2018;52:103-9. [Crossref] [PubMed]
- Liu WJ, Du Y, Wen R, et al. Drug resistance to targeted therapeutic strategies in non-small cell lung cancer. Pharmacol Ther 2020;206:107438. [Crossref] [PubMed]
- Gou LY, Niu FY, Wu YL, et al. Differences in driver genes between smoking-related and non-smoking-related lung cancer in the Chinese population. Cancer 2015;121:3069-79. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Zhou J, Zhao C, Zhao J, et al. Re-biopsy and liquid biopsy for patients with non-small cell lung cancer after EGFR-tyrosine kinase inhibitor failure. Thorac Cancer 2019;10:957-65. [Crossref] [PubMed]
- Tan DS, Camilleri-Broët S, Tan EH, et al. Intertumor heterogeneity of non-small-cell lung carcinomas revealed by multiplexed mutation profiling and integrative genomics. Int J Cancer 2014;135:1092-100. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Turner NC, Reis-Filho JS. Genetic heterogeneity and cancer drug resistance. Lancet Oncol 2012;13:e178-85. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov 2021;11:858-73. [Crossref] [PubMed]
- Lebofsky R, Decraene C, Bernard V, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015;9:783-90. [Crossref] [PubMed]
- Guo Q, Wang J, Xiao J, et al. Heterogeneous mutation pattern in tumor tissue and circulating tumor DNA warrants parallel NGS panel testing. Mol Cancer 2018;17:131. [Crossref] [PubMed]
- Piotrowska Z, Sequist LV. Epidermal Growth Factor Receptor-Mutant Lung Cancer: New Drugs, New Resistance Mechanisms, and Future Treatment Options. Cancer J 2015;21:371-7. [Crossref] [PubMed]
- Koldby KM, Mortensen MB, Detlefsen S, et al. Tumor-specific genetic aberrations in cell-free DNA of gastroesophageal cancer patients. J Gastroenterol 2019;54:108-21. [Crossref] [PubMed]
- McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15-26. [Crossref] [PubMed]
- Jiang T, Ren S, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer 2015;90:128-34. [Crossref] [PubMed]
- Leers MPG. Circulating tumor DNA and their added value in molecular oncology. Clin Chem Lab Med 2020;58:152-61. [Crossref] [PubMed]
- Yu L, Liu X, Han C, et al. XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of hepatitis B virus-related hepatocellular carcinoma after hepatectomy: A cohort study for 10 years' follow up. Hepatol Res 2016;46:765-74. [Crossref] [PubMed]
- Huang A, Zhao X, Yang XR, et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol 2017;67:293-301. [Crossref] [PubMed]
- Cai ZX, Chen G, Zeng YY, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer 2017;141:977-85. [Crossref] [PubMed]
- Huang A, Zhang X, Zhou SL, et al. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer 2016;7:1907-14. [Crossref] [PubMed]
- Gormally E, Vineis P, Matullo G, et al. TP53 and KRAS2 mutations in plasma DNA of healthy subjects and subsequent cancer occurrence: a prospective study. Cancer Res 2006;66:6871-6. [Crossref] [PubMed]
- Lim M, Kim CJ, Sunkara V, et al. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (CTCs and ctDNA). Micromachines (Basel) 2018;9:100. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Panagopoulou M, Karaglani M, Balgkouranidou I, et al. Circulating cell-free DNA in breast cancer: size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2019;38:3387-401. [Crossref] [PubMed]
- Ma M, Zhu H, Zhang C, et al. "Liquid biopsy"-ctDNA detection with great potential and challenges. Ann Transl Med 2015;3:235. [PubMed]
- Namløs HM, Boye K, Meza-Zepeda LA. Cell-free DNA in blood as a noninvasive insight into the sarcoma genome. Mol Aspects Med 2020;72:100827. [Crossref] [PubMed]
- Gautschi O, Huegli B, Ziegler A, et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett 2007;254:265-73. [Crossref] [PubMed]
- Parkinson CA, Gale D, Piskorz AM, et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med 2016;13:e1002198. [Crossref] [PubMed]
- Morikawa A, Hayashi T, Shimizu N, et al. PIK3CA and KRAS mutations in cell free circulating DNA are useful markers for monitoring ovarian clear cell carcinoma. Oncotarget 2018;9:15266-74. [Crossref] [PubMed]
- Loft M, Lee B, Tie J, et al. Clinical Applications of Circulating Tumour DNA in Pancreatic Adenocarcinoma. J Pers Med 2019;9:37. [Crossref] [PubMed]
- Chen H, Tu H, Meng ZQ, et al. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol 2010;36:657-62. [Crossref] [PubMed]
- Däbritz J, Preston R, Hänfler J, et al. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas 2009;38:534-41. [Crossref] [PubMed]
- Trojan J, Klein-Scory S, Koch C, et al. Clinical Application of Liquid Biopsy in Targeted Therapy of Metastatic Colorectal Cancer. Case Rep Oncol Med 2017;2017:6139634. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


