
Protein kinase CK2 in DNA damage and repair
Introduction
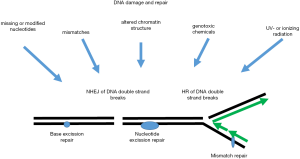
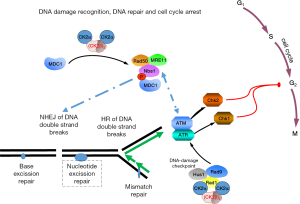
Human chromosomes are constantly under pressure from various DNA damaging agents, such as ultraviolet light (UV), reactive oxygen species, ionizing radiation and a lot of different chemicals in the environment (1). These various insults create DNA single or double strand breaks and various modifications of bases in the nucleotides. Furthermore, there are defects generated naturally by thermally induced alterations in purine- and pyrimidine bases as well as mutations induced by DNA replication (Figure 1). The cellular response to DNA damage includes three general steps i.e., (I) recognition of DNA damage; (II) intra-cellular signalling leading to cell cycle arrest; (III) DNA repair. DNA damage is recognized by various sensors, which then usually induce a variety of cellular signalling pathways including pathways that regulate cell cycle arrest giving the cell time to repair the damage. Depending on the DNA damage various repair mechanisms are known such as base excision repair, nucleotide excision repair, mismatch repair, homologous and non-homologous end joining, just to mention but a few (2,3) (Figure 1). Among the many proteins which are implicated in the recognition of DNA damage and the following cellular signalling are protein kinases such as the phosphatidylinositol-like kinases ATM, ATR and DNA-PK (1,3). These and many other kinases phosphorylate a variety of down-stream effectors many of which are recruited to the DNA damage, are somehow implicated in cell cycle arrest or are necessary for DNA repair. Mutations in the genes coding for these kinases result in embryonic lethality or strong cancer predisposition. Among the human kinome over the last 10 years protein kinase CK2 has attracted the attention of many scientist because this enzyme seems to be required for cell viability (4). The amount and the enzyme activity of CK2 are elevated in cancer cells (5). Furthermore, more and more inhibitors of the kinase activity were detected which are partially useful drugs for the treatment of cancer (6,7). In the present review I will focus on the role of protein kinase CK2 in DNA damage recognition, subsequent signalling and DNA repair.
More than 60 years after the first description of its enzymatic activity protein kinase CK2 is still an enigma. It is implicated in a variety of different cellular processes such as regulation of cell proliferation, apoptosis, signal transduction, development and differentiation, regulation of transcription, angiogenesis, carbohydrate metabolism and protein biosynthesis just to mention but a few (4,8-12). There is a steadily increasing number of different substrates in nearly every compartment of a eukaryotic cell (13,14). Thus, CK2 seems not to be implicated in the regulation of one particular single pathway but it seems to modulate many different pathways depending on the subcellular localisation, interacting proteins and the cell species (14,15). CK2 is composed of two catalytic α- or α'- subunits which are encoded by distinct genes on different chromosomes (16) and two non-catalytic β-subunits to form the CK2 holoenzyme. In addition, there is increasing evidence for functions of the individual CK2 subunits aside from the holoenzyme (17) (and literature therein) and moreover, there is also some evidence that the activity of CK2 is regulated by aggregation into higher molecular complexes. Although primarily classified as a serine/threonine kinase, CK2 also phosphorylates tyrosine residues (18-20).
Since CK2 is for a long time known to be implicated in so many cellular processes such as regulation of life and death of a cell, cell proliferation and cell cycle regulation it was not surprising that already 20 years ago the first reports appeared demonstrating also a role in genotoxic stress response.
CK2 influence on chromatin structure
Post-translational modifications including phosphorylation of histones are known to modify the histone-DNA interactions. CK2 phosphorylates histone H4 at serine 1 and it was further shown that this phosphorylation slightly affected the efficiency of DNA double-strand break repair by non-homologous end joining (21,22). On the other hand a histone H4 serine 1 mutant did not show any elevated sensitivity to DNA damage. Instead, it was shown that an elevated serine 1 phosphorylation by CK2 correlates with histone de-acetylation (22).
Shortly after DNA damage histone H2AX is phosphorylated and this phosphorylation is an early known marker of DNA damage. CK2 was found to co-localize with phospho H2AX (γH2AX) at sites of DNA strand breaks. Down-regulation of CK2α and CK2α' by small interfering RNAs (siRNAs) and treatment of the cells with a radiomimetic compound revealed a higher level of γH2AX which might indicate a higher level of DNA strand breaks (23). Proximity ligation assays (PLA assays) showed a co-localization of γH2AX and CK2α' demonstrating one of the very rare functions of CK2α'. Phosphorylated γH2AX recruits other proteins to the site of DNA damage in order to activate DNA repair. Another protein which is recruited to sites of DNA damage is the heterochromatin protein HP1-β also named CBX1. This protein is bound to histone H3 which is methylated at lysine 9. CK2 phosphorylation seems to be implicated in HP1-β mobilization and γH2AX phosphorylation. Thus, these data point to a CK2 mediated signalling mechanism that initiates the DNA damage response by altering the chromatin structure (24,25).
Recently, it was shown that the number of DNA single strand breaks did not depend on pre-treatment of the cells with a CK2 kinase inhibitor, namely TBB (26). This might indicate that CK2 does not play a significant role for DNA damage induction. Instead it was found that the disappearance of foci was delayed after inhibition of the CK2 kinase activity as measured by γH2AX focus formation. The authors speculated that CK2 might regulate the γH2AX dephosphorylation by its ability to regulate the PP2A phosphatase (27). Alternatively, CK2 phosphorylation of HP1-β in response to DNA damage might play a role in the regulation of γH2AX phosphorylation (24).
CK2 and cellular signalling
The first indication for a role of CK2 in the cellular response to DNA damage stems from an experiment described in 1990 where a cDNA coding for CK2α complemented the UV-sensitivity of an immortalized Xeroderma pigmentosum cell line to an UV resistant level (28). Since transfection of CK2α into these cells resulted in an increase in CK2 protein kinase activity the authors speculated about the possibility of a cellular response to DNA damage by CK2 dependent protein phosphorylation. The first substrate which was identified as a CK2 substrate and CK2 binding partner implicated at least indirectly in DNA repair was the growth suppressor p53Jeny (29-32). Interestingly, binding between CK2 and p53 was mediated by the β-subunit of CK2 which led to a down-regulation of the p53 transactivation function. On the other hand, binding of wild-type p53 to CK2 down-regulated the CK2 kinase activity whereas a transforming mutant of p53 enhanced CK2 kinase activity (31-34). p53 was found to promote the rapid annealing of complementary RNA and DNA strands, whereas pre- incubation of p53 with CK2 completely inhibited the p53 annealing activity (32). In other experiments it was shown that arsenite inhibited an UVB induced phosphorylation of human p53 at the CK2 phosphorylation site serine 392 (35). This inhibition corresponded to a suppression of the p53 DNA binding activity and transactivation function. The authors speculated that arsenite may act as a co-carcinogen by targeting p53 thus, inhibiting DNA repair. Other DNA damage inducing factors such as psoralen and UVA also modify human p53 at amino acid 392Jeny (36-39). Mutation of serine 389, which is the corresponding CK2 phosphorylation site in mouse p53, led to enhanced UV-induced skin cancer which indicates a tumour-suppressing role of p53 in UV-induced DNA damage (40). Keller et al. identified an UV-activated protein complex which contained CK2 and the chromatin elongation factor FACT (facilitates chromatin transcription). FACT is a heterodimer containing hSpt16/cdc68 and SSRP1 (structure-specific recognition protein 1) (41). This UV-activated protein complex specifically phosphorylated serine 392 of human p53 and thereby enhanced the sequence- specific DNA binding of p53 at least in vitro (42,43). The enhanced DNA binding activity correlated with an elevated transcriptional activity of p53 in response to UV. Later on maize SSRP1 was identified as a substrate for CK2 (44). This phosphorylation induced the recognition of UV-damaged DNA.
Recognition of the DNA damage induces a signalling cascade which is started by the PI3-kinase-like kinases ATM, ATR and DNA-PK followed by the down-stream checkpoint effector kinases Chk1, Chk2 and Chk3 (MK2). One of the cell cycle regulating kinases and down-stream targets of Chk1 and Chk2 is cdk1 which is phosphorylated by CK2 at amino acid serine 39 (45). Furthermore, phosphorylation of other substrates by these PI3-kinase-like kinases leads to the formation of multi-protein complexes at the damaged DNA to link DNA damages to cell cycle arrest (46).
Ataxia-telangiectasia mutated (ATM) protein is a protein kinase which is activated in response to DNA damage. One of the targets of this kinase is the p53-binding protein 1 (53BP1). This protein is characterized by BRCA1 C-terminal domains which are implicated in protein-protein interactions, Tudor domains for the recruitment to the chromatin and a glycine-arginine-rich (GAR) motif which is required for DNA binding. There is ample evidence that 53BP1 contributes to DNA damage repair and suppression of genomic instability (47,48). Knock-down by siRNA experiments revealed that CK2 seems to be necessary for ATM and Chk2 phosphorylation as well as for a 53BP1 foci formation (49). By co- immunoprecipitation experiments it was further shown that CK2 bound to 53BP1 through its GAR and Tudor domains. This complex formation is impaired in the presence of DNA damage.
A more direct role of CK2 in DNA repair was found when the human DNA repair protein apurinic/apyrimidinic endonuclease (APE) was identified as a CK2 substrate (50). It was shown that phosphorylation of APE by CK2 resulted in a total inactivation of the APE activity. A direct link to p53 was detected when it was shown that APE converts inactive p53 into an active form (51). In a later report, phosphorylation of APE by CK2 was confirmed, however, there was no difference in APE endonuclease activity for CK2 phosphorylated APE (52). Instead, the authors found that the association between the transcription factor AP-1 and APE was inhibited when APE had been phosphorylated by CK2. Based on experiments with an inhibitor of the CK2 kinase activity, namely quercetin, the authors speculated that inhibition of the CK2 phosphorylation of APE might contribute to an increased sensitivity to DNA damage.
It is known for quite some time, that the tumour suppressor gene product BRCA1 participates in DNA repair (53). BRCA1 gets phosphorylated after UV treatment of normal keratinocytes in a dose-dependent manner. Bicyclic monoterpene diols (BMT diols) to some extent can mimic UV treatment of cells. Thus, it was shown that the BMT diol 2,2-dimethyl-3-propanyldiol-norbonane induced phosphorylation of BRCA1 and stimulated the repair of UV-induced pyrimidine dimers (54). Since it was already shown that BRCA1 is a substrate for CK2 (55) inhibition experiments of CK2 with DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) (56) revealed a complete inhibition of BRCA1 phosphorylation (54). This inhibition of the BRCA1 phosphorylation is accompanied by an inhibition of the BMT diol stimulated repair of thymidine dimers.
Rad52 from yeast and mammals are key components of DNA double-strand break repair. Sumoylation of Rad52 is induced by DNA damage. This post-translational modification regulated Rad52 stability and activity (57). Rad52 specifically interacted with PTEN and this interaction in particular in the nucleus was enhanced after DNA damage (58) as detected by co-immunoprecipitation experiments as well as by co-immunofluorescence studies. Furthermore, CK2 phosphorylation of PTEN led to nuclear translocation of PTEN where it interacted with Rad52 thereby regulating the sumoylation of Rad52 (58). In eukaryotes Rad51 together with Rad52 and Rad54 catalysed strand transfer between the damaged sequences and its undamaged homologue to allow re-synthesis of the damaged region.
CK2 and mismatch repair
Cells are in constant need for an efficient DNA repair, because up to 104 spontaneous depurinations take place in one single cell per day (59). One of the repair mechanisms is mismatch repair, which plays an important role in the maintenance of genomic stability by removal of mispaired nucleotides from DNA. Both mismatch binding proteins MSH2 and MSH6, which form the MutSα complex, are substrates for protein kinase CK2 (Figure 2). Inhibition of CK2 by quercetin resulted in a reduced binding of MSH2/MSH6 to mismatches. This result was confirmed by dephosphorylation experiment for MSH2/MSH6 thus providing the first indication for a role of CK2 in mismatch repair (21).
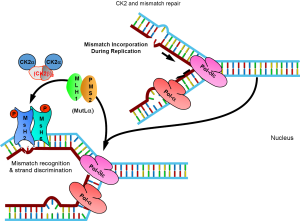
The nucleotide analogue 6-thioguanine is used as an efficient drug in the chemotherapy of a variety of tumours (60). This nucleotide analogue is incorporated into DNA which may later result in an increase in DNA single strand breaks. Incorporation of 6-thioguanine also induces mismatch repair. Following treatment of HeLa cells with 6-thioguanine, CK2α re-localized from the nucleus to the endoplasmic reticulum. No re-localization was found for CK2α' (45). Further evidence for a role of CK2 in mismatch repair stems from experiments which showed that in vitro phosphorylation of cell extracts by CK2 increased the cleavage of 8-oxoguanine oligonucleotide considerably (61). 8-Oxoguanine is generated by oxidative stress. It readily forms mismatches with cytosine or adenine. Mutγ is responsible for the repair of 8-oxoguanine. It was not clear, however, whether CK2 phosphorylation of Mutγ is directly implicated in DNA repair or whether it regulates intracellular localization or protein-protein interaction with other repair proteins.
CK2 and nucleotide excision repair
Nucleotide excision repair (NER) is one of the most versatile mammalian DNA repair mechanisms. The transcription factor TFIIH complex Xeroderma pigmentosum group B (XPB) helicase subunit not only functions in transcription but also in NER. XPB was phosphorylated by CK2 in the C-terminus (62). This phosphorylation led to an impaired NER whereas the helicase activity of XPB was not affected. The explanation for this observation might be that CK2 phosphorylation of XPB regulates the recruitment or positioning of other factors which are inevitable for DNA repair. Some years ago it was shown that another component of the TFIIH complex namely cyclin H is a substrate for CK2 (63) and CK2 phosphorylation regulates cyclin H/cdk7/Mat1 activity. Thus, at least two components of the NER TFIIH complex are CK2 substrates.
Centrins are Ca2+ binding proteins, which interact with the Xeroderma pigmentosum group C (XPC) protein, which is also implicated in nucleotide excision repair (64). Phosphorylation of centrins by CK2 regulated binding to G-proteins (65) in a light dependent fashion. CK2 phosphorylation of centrin1 weakly reduced its binding to XPC whereas CK2 phosphorylation of centrin2 reduced binding to XPC considerably (66). The molecular consequences of this reduced binding of centrin2 to XPC remains to be elucidated.
CK2 and non-homologous end joining and homologous recombination
XRCC1 dependent processes
The X-ray repair cross-complementing group 1 (XRCC1) is a member of a family of XRCC proteins which play important roles in these types of DNA repair (67). XRCC1 deficient cells show an accumulation of DNA single-strand breaks after DNA damage and elevated levels of chromosomal aberrations. XRCC1 binds directly to DNA single-strand breaks and gaps in DNA. Since no enzymatic activity was found for XRCC1 it was suggested that XRCC1 is a platform for the binding of other proteins at places of DNA damage.
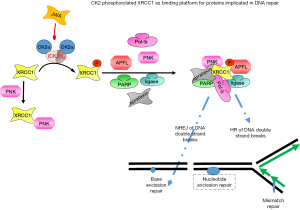
XRCC1 is directly implicated in the repair of DNA single-strand breaks after exposure of DNA to ionizing radiation or alkylating agents. It interacts with poly (ADP-ribose) polymerase (PARP) and DNA ligase to participate in base excision repair and homologous recombination (68) (Figure 3). In the presence of CK2 the DNA ligase I activity was reduced in a CK2 dose-dependent manner (69). This might be due to the phosphorylation of DNA ligase I at serine 66 (35,70). Since DNA ligase I participates in homologous DNA repair and nucleotide excision repair these data support the idea about the important role of CK2 in DNA damage repair. Phosphorylation of XRCC1 by CK2 led to an elevated stability of DNA ligase III (71). XRCC1 is phosphorylated by CK2 in vitro and in vivo (54,72,73). These initial findings were recently extended by the observation that the CK2 phosphorylated XRCC1 interacts with different regions of the fork head associated domain of PNK while the non-phosphorylated XRCC1 binds to the catalytic domain of PNK with lower affinity (50,61,73). Human polynucleotide kinase (PNK) is a DNA repair enzyme which modifies DNA termini to prepare them for DNA repair. It was shown that CK2 phosphorylation of XRCC1 is required for the assembly of XRCC1 into foci at DNA strand breaks together with PNK (73). A role for CK2 in the phosphorylation of XRCC1 was also demonstrated by studies with a CK2 inhibitor (54) and by siRNA knock-down experiments (74). It was recently demonstrated that the cytoplasmic variant of CK2 phosphorylated XRCC1 (71) and this phosphorylation inhibited ubiquitination and proteasomal degradation of XRCC1 as shown by siRNA knock-down of CK2α and CK2α' as well as by mutation of the CK2 phosphorylation sites on XRCC1.
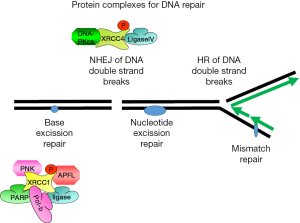
Aprataxin is a protein with some homology to PNK which also plays a role in DNA repair by binding to XRCC1 (53,54). Aprataxin only binds to CK2 phosphorylated XRCC1. Furthermore, binding of aprataxin to XRCC1 was shown to be essential for maintaining the steady-state level of XRCC1. In addition to PNK and aprataxin a third protein denoted aprataxin—and PNK-like factor (APLF) which is also known as Xip1 or PALF (38) were found to bind to XRCC1 and X-ray repair cross-complementing protein 4 (XRCC4) in a CK2 phosphorylation-dependent manner (69). Depletion of APLF is associated with impaired non-homologous end joining (NHEJ) (75). APLF possesses endonuclease and 3'-5' exonuclease activities, it accumulates at sites of single-strand breaks or double-strand breaks where it seems to act as a DNA end-processor. Binding of XRCC1 to APLF seems to be necessary for the regulation of the steady state of APLF. The importance of APLF for DNA damage recognition was demonstrated by depletion experiments which led to a significantly reduced survival in response to single-strand breaks. These results suggest that XRCC1 can recruit one of the three different factors depending on the type of DNA damage. Altogether, phosphorylation of XRCC proteins by CK2 generates a platform on the XRCC1 polypeptide chain where a number of different proteins with functions in DNA damage recognition and repair can bind to.
It was further shown that the phosphorylation of XRCC1 by CK2 was responsible for the translocation of XRCC1 to the nuclear matrix upon oxidative DNA damage by H2O2 (76). Thus, another possible role for CK2 might be the direction of repair proteins to places of rapid DNA repair response.
As mentioned earlier, phosphorylation of XRCC1 by CK2 is known to stimulate the repair of single-strand breaks (71,73). Expression of a variant of XRCC1 which cannot be phosphorylated by CK2 leads to low single-strand breaks after treating the cells with the alkylating agent dimethyl sulfate whereas XRCC1 deficient cells displayed elevated levels of single-strand breaks. This reduction in the amount of single-strand breaks was also obtained by using the CK2 specific inhibitor 4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) (77). The reduction of single-strand breaks in the presence of the mutant XRCC1ckm is accompanied by fewer RAD51 foci following dimethyl sulfate treatment of XRCC1ckm cells. Interestingly, most of the non-CK2 phosphorylatable XRCC1ckm was found in a chromatin bound fraction whereas only 25% were found in the soluble fraction.
Induction of DNA damages is a versatile strategy for the treatment of cancers in humans. One of the commonly used cytostatic drugs for the treatment of cancer is cisplatin which induces DNA damage through crosslinking of DNA. XRCC1 was found to bind to such cisplatin induced crosslinks (78). It was recently shown that the selective CK2 inhibitor CX-4945, which is in stage II clinical trials, blocks DNA repair after cisplatin or gemcitabine induced DNA damages. This block in DNA repair is due to an inactivation of XRCC1 and MDC1 (79,80). These data support the idea that CK2 inhibition is a strategy for a combination therapy of cancer treatment.
JWA is a growth suppressor with multiple functions in cellular responses to oxidative stress and base excision repair as well as melanoma cell adhesion, invasion and metastasis (81-83). A recent study provided data showing that JWA negatively regulated XRCC1 in cisplatin-resistant cells (Figure 3). It enhanced cisplatin induced cell death by upregulation of XRCC1 in cisplatin-sensitive cells (80). It was further shown that overexpression of JWA inhibits the CK2 mediated phosphorylation of XRCC1 which is supported by the fact that mutation of the CK2 phosphorylation residues on XRCC1 blocks the negative regulation of JWA on XRCC1.
XRCC4 dependent processes
Very similar observations to XRCC1 were described for the XRCC4, which is one of the several proteins involved in the non-homologous end joining (NHEJ) pathway to repair DNA double strand breaks (DSBs) (Figure 4). XRCC4 is a substrate for CK2 and CK2 phosphorylation of XRCC4 is necessary for its interaction with PNK. This interaction is important for DNA double-strand break repair (61) demonstrating that CK2 regulates not only single-strand break repair but also double-strand break repair. In both cases the scaffold proteins XRCC1 and XRCC4 are the link between DNA binding and protein-protein interaction between XRCCs and various proteins. These interactions ensure that DNA end processing and DNA ligation are coordinated. XRCC4 directly interacts with DNA ligase IV, thereby stimulating the ligase activity (84). The XRCC4/DNA ligase IV complex is recruited to DNA double-strand breaks by the DNA dependent kinase (DNA-PK).
Ku70/Ku80/DNA-PK dependent processes
XRCC5 also known as Ku80 and XRCC6 known as Ku70 are components of the DNA-dependent protein kinase complex (DNA-PK) where XRCC7 represents the catalytic subunit of DNA-PK (known as DNA-PKcs). The Ku80 and Ku70 proteins form heterodimers which bind to DNA ends acting as a sensor for DNA damage. In non-homologous end joining (NHEJ) DNA-PK plays a role in the detection of DNA double-strand breaks and in the ligation of the DNA ends. Binding of Ku-proteins to double-strand breaks serves to recruit the catalytic subunits of DNA-dependent protein kinase (DNA-PKcs, ligases and DNA polymerase (85). DNA-PK is a multiply phosphorylated protein. Down-regulation of CK2 subunits by siRNA experiments affected the activity and auto phosphorylation status of DNA-PK which resulted in an inhibition of DNA-PK dependent DNA double-strand break repair. No phosphorylation of DNA-PK by CK2 was shown, however, a proximity ligation assay clearly revealed an interaction of CK2 subunits with DNA-PK. Moreover, as shown by co-immunoprecipitation experiments CK2α and CK2α' bound to DNA-PKcs whereas no binding was found for CK2 and Ku80. Down-regulation of CK2α and CK2α' led to a decrease in the auto-phosphorylation of DNA-PKcs (86). Further experiments showed that the CK2 kinase activity seems to be dispensable for the DNA-PKcs auto-phosphorylation (23). The association between DNA-PKcs and CK2 increased upon induction of DNA strand breaks. In summary a lack of CK2α and CK2α' leads to persistent DNA damage and finally to a reduced survival rate in cells treated with a radiomimetic compound. Probably this interaction is necessary for the phosphorylation of Ku by CK2 (86).
It is an interesting observation that a lack of DNA-PKcs was shown to be accompanied by an increase in the mRNA and protein level for CK2α' but not for the other CK2 subunits. It was further shown that there was no DNA amplification for CK2α' (87) indicating that DNA-PKcs regulated the expression of CK2α'.
Both Ku70 and Ku80 bind to inositol hexakisphosphate (IP6) and this interaction seems to be necessary for NHEJ. Biotinylated IP6 was used to precipitate IP6 binding proteins. This approach demonstrated CK2 as one of the IP6 binding proteins, which is in agreement with earlier results showing that CK2 bound to IP6. It was also shown that IP6 regulates CK2 activity by replacing the CK2 inhibitor NOPP140 from CK2. With these results IP6 was identified as an activator of CK2 (88,89). One might speculate that the Ku70/Ku80 associated IP6 modulates the activity of CK2 for NHEJ.
Mre11-Rad50-Nbs1 dependent processes
The Mre11-Rad50-Nbs1 complex also known as MRN complex has emerged as one of the main complexes for homologous recombination as well as for non-homologous end joining. The MRN complex provides the platform for the detection of DNA breaks, it activates DNA repair mechanisms and processes DNA ends for the final ligation process. On the other hand the MRN complex activates the ATM kinase thus linking DNA repair to cell cycle regulation (90). Similar to Mre11, a member of the ABC superfamily of ATPases, Nbs1 forms a dimer within the MRN complex. A function of Nbs1 seems to be the binding platform for a number of different proteins such as CtIP, MDC1, ATR and WRN. It turned out that MDC1 is phosphorylated by CK2 and this phosphorylation seems to be required for its interaction with the MRN complex (91,92) and for the accumulation at DNA double-strand breaks (Figure 5). The MDC1 interaction with the MRN complex resembles the interaction previously shown for XRCCs and PNK, aprataxin or APLF. Although the interaction between XRCCs and PNK, aprataxin or APLF is sensitive to pharmacological inhibition of CK2 the MRN-MDC1 interaction is mostly unaffected. A step forward in understanding how the MRN complex and in particular Nbs1 within the MRN complex contributes to DNA repair came from the identification of TCOF1, a nuclear factor, which transiently co-localizes with Nbs1 in the nucleolus rom after DNA damage. Experiments with siRNAs for CK2α and CK2α' revealed that these two subunits were required for the recruitment of Nbs1 to the nucleolus. TCOF1 was phosphorylated by CK2 (93) and it was proposed that MDC1 and TCOF1 interact with Nbs1 in a similar manner through the binding of CK2 sites to Nbs1. This assumption would mean that binding of TCOF1 and MDC1 to Nbs1 is mutually exclusive.
The N-terminus of MDC1 contains many putative CK2 phosphorylation sites. Among these various sites are at least six SDTD motifs within an acidic environment. Interestingly, Nbs1 interacts only with doubly phosphorylated SDTD motifs (22,92). Aprataxin is known to be defective in the neurodegenerative disorder ataxia oculomotor apraxia type 1. This protein binds to sites of DNA damage by an interaction with CK2 phosphorylated SDTD motifs on the polypeptide chain of MDC1 (94). Mutational analysis, however, revealed that CK2 phosphorylation is not the sole factor required for this interaction because mutation of arginine 29 disrupted the interaction between aprataxin and MDC1. Further experiments have shown that MDC1 is not necessary for the recruitment of aprataxin to sites of DNA strand breaks but rather MDC1 seems to be a platform for aprataxin and probably other proteins to ensure an effective DNA repair. Interestingly, CK2 seems not to be located at sites of DNA damage, which means that CK2 phosphorylation occurs before the association of these proteins with damaged DNA.
In Schizosaccharomyces pombe the Mre11-Rad50-Nbs1 complex also plays an important role in homologous recombination. The Ctp1 protein cooperates in this process and it is recruited to DNA single strand breaks by interacting with the MRN complex. CK2 phosphorylated SDTD motifs serve as the preferred docking sites for Ctp1/Nbs1 binding. Thus, it has been shown that the CK2 phosphorylation dependent protein-protein interaction not only plays a role in non-homologous end joining (NHEJ), single-strand break repair but also in homologous repair (53,75,95).
One other factor which is implicated in NHEJ is the Rad51 recombinase, which catalyses homologous pairing and strand exchange during homologous recombination. Recruitment and activity of Rad51 is among others stimulated by BRCA2. Rad51 is phosphorylated by CK2. This phosphorylation is primed by a previous phosphorylation by polo like kinase 1 (Plk1). It was further shown that the combined phosphorylation of Rad51 by Plk1 and CK2 triggers its interaction with Nbs1. CK2 phosphorylation of Rad51 is important for the co-ordination of precise recombination events at the DNA (96).
Besides the MRN complex, MDC1 and 53BP1, the MDC1 dependent recruitment of E3 ligase RNF8-RNF168 facilitates DNA strand break signalling and DNA repair. The recruitment of 53BP1 to DNA damage sites seems to depend on two histone modification such as methylation of H4K20 and ubiquitination of H2AK15. RNF168 is required for the recruitment of the lysine-specific demethylase LSD1 to sites of DNA damage. By co-immunoprecipitation assays and by pull-down assays it was shown that LSD1 bound to CK2α' and to a lesser extent to CK2α. In addition LSD1 was phosphorylated by CK2α' (97). This is one of the very few reports indicating a specific role of CK2α'.
Miscellaneous CK2/protein interactions
TopBP1 is a multifunctional protein, which is has been identified as a binding partner of DNA topoisomerase II (98,99). Later on, a member of the TopBP family of proteins, namely TopBP1, turned out to be a scaffold for numerous protein-protein interactions through BRCT (BRCA1 C-terminus) domains. TopBP1 binds to sites of DNA damage and to Rad9 within the Rad9-Hus1-Rad1 (9-1-1) checkpoint clamp. Moreover, serine 387 of Rad9 which is required for its interaction with TopBP1 is phosphorylated by CK2 (100). In addition TopBP1 stimulates the kinase activity of ATR. Thus, the CK2 substrate TopBP1 seems to be the link between DNA repair and cell cycle regulation. In the same year Takeishi et al., reported that in addition to serine 387 also serine 341 was phosphorylated by CK2 at least in vitro (101). TopBP1 bridges the 9-1-1 checkpoint clamp and the ATM-ATR interacting protein which are recruited independently onto damaged chromatin. Recently, it was shown that the recruitment of Rad9 to UV-damaged DNA is independent of its binding to TopBP1 (102). The Rad9-Hus1-Rad1 complex is a heterotrimer which is loaded onto damaged chromatin (103). It was demonstrated that CK2 phosphorylates Rad9 within the C-terminal tail (101). The phosphorylated serine residue 387 has been shown to be important for binding to TopBP1. It turned out that in addition to serine 387 also CK2 phosphorylation at residue 341 seems to be important for DNA damage response in human cells. Phosphorylation at these sites are not DNA-damage dependent. This observations seems to be very similar to a previous result with MDC1 which is also constitutively phosphorylated by CK2 and the interaction between MDC1 and Nbs1 takes place in the absence of DNA damage (91,92). Furthermore, by tandem mass spectrometry and by co-immunoprecipitation experiments it was shown that CK2α, CK2α' and CK2β interacted with the Rad9-Hus1-Rad1 complex (Figure 5).
Shortly after ionizing radiation of malignant glioma cells the catalytic activity of CK2 increased and this activity was inhibited by apigenin (104) a flavonoid, which was used in a clinical phase II study for the treatment of colorectal cancer (NCT 00609310). It was reported that UV treatment of tumour cells resulted in enhanced expression of CK2α (105,106). It turned out that CK2 knock-down in the malignant glioma cells did not inhibit DNA double strand break repair (104). It was further shown that apigenin increased the radiation induced phosphorylation of CK2, which is a target of DNA-PK (107). CK2 inhibition by apigenin did not radio-sensitize the two tested glioma cell lines. Inhibition of CK2 by tetrabromobenzotriazole (TBB) or down-regulation of CK2 by siRNA experiments increased apoptosis in human lung cancer cells, which is accompanied by elevated cytochrome C release from mitochondria and by cleaved caspase 3.
HspA1A also known as Hsp70 is a member of the family of inducible heat shock proteins in mammalian cells, which protects cells against DNA damage (108). HSPA1A binds to apurinic/apyrimidinic endonucleases (109) to stimulate endonuclease activity. It binds to XRCC1 (109) and it enhances base excision repair after ionizing radiation (110). Recently, by co-immunoprecipitation it was found that HSPA1A bound to CK2 (111) and this interaction increased after treatment of the cell with the DNA damaging agent benzo[a]pyrene. Con-focal immunofluorescence microscopy revealed that HSPA1A and CK2 co-localized in the cell nucleus and perinuclear. Furthermore, it was shown that overexpression of HSPA1A resulted in an enhanced CK2 kinase activity. It might well be that the interaction between HSPA1A and CK2 targets CK2 to sites of DNA damage where CK2 is activated by HSPA1A to phosphorylate XRCC1 or DNA-PK.
Another protein which is implicated in DNA damage repair is OTUB1 which is a member of isopeptidases, which remove ubiquitin chains form proteins (112,113). It was recently shown that CK2α phosphorylates OTUB1 in vitro and in vivo. Further experiments revealed that OTUB1 belongs to CK2 substrates, which are phosphorylated by the catalytic CK2α subunit alone as well as by the CK2 holoenzyme. CK2 phosphorylation of OTUB1 at serine 16 promotes nuclear localisation of OTUB1 (114). Furthermore, CK2 phosphorylation of OTUB1 seems to be necessary for the repair of IR induced DNA damage. The precise role of OTUB1 in DNA repair, however, is not resolved.
Concluding remarks and perspectives
Protein kinase CK2 is a multifunctional enzyme which plays a role in diverse cellular processes. Here, I have added functions of CK2 in DNA damage recognition and repair and down-stream signalling. Early reports on CK2 in this context pointed already to a role of CK2 in the regulation of the chromatin structure, which was evident by the CK2 phosphorylation of histones. One of the key molecules in sensing DNA damage, induction of cell cycle arrest and DNA repair is p53, which was detected as a CK2 substrates in the early 90 of the last century. Components of the transcription factor complex TFIIH, which is also known to be implicated in DNA repair, were also identified as CK2 substrates (Table 1). In the course of these studies there were the first indications that CK2 not only phosphorylated various substrates in the context of DNA damage and repair but individual subunits of CK2 or the holoenzyme bound to cellular proteins where the functional consequences of this binding remains to be elucidated. Binding of CK2 to various proteins might in many cases reflect an enzyme/substrate interaction. There is, however, also the possibility that CK2 is targeted to specific substrates by binding to transport proteins.
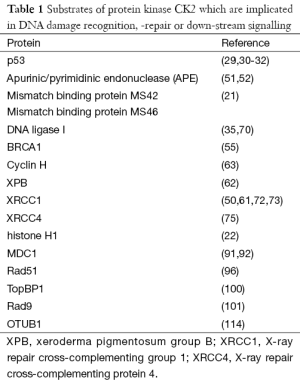
Full table
CK2 also links DNA damage recognition to cell cycle arrest, which gives the cell time for DNA repair. This is mostly evident by the observation that CK2 seems to be necessary for p53, p21WAF1, ATM, ATR, Chk2 and cdk1 phosphorylation.
CK2 is implicated in the phosphorylation of proteins, which are central in mismatch repair, nucleotide excision repair, homologous recombination and non-homologous end joining. Interestingly CK2 phosphorylates proteins such as XRCC1, XRCC4 and MDC1, which are platform proteins for the recruitment of other proteins to the damaged DNA for repair. CK2 phosphorylation of these proteins regulates binding of these proteins to the platform proteins.
Over the last 10 years a number of inhibitors of the kinase activity have been developed and a few of them are in clinical phase I or II trials. On the other hand, the induction of DNA damage by various cytostatic is a strategy for the treatment of cancer. Actually there are a few studies coming up where combinations of CK2 inhibitors and well known DNA damage inducing cytostatic have been used for the treatment of cancer with promising results. These initial results open a wide field for future research about CK2 in clinical trials.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2016.01.09). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Roos WP, Kaina B. DNA damage-induced cell death:from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 2013;332:237-48. [PubMed]
- Reyes GX, Schmidt TT, Kolodner RD, et al. New insights into the mechanism of DNA mismatch repair. Chromosoma 2015;124:443-62. [PubMed]
- Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 2016;16:20-33. [PubMed]
- St-Denis NA, Litchfield DW. From birth to death:The role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell Mol Life Sci 2009;66:1817-29. [PubMed]
- Trembley JH, Wang G, Unger G, et al. CK2:A key player in cancer biology. Cell Mol Life Sci 2009;66:1858-67. [PubMed]
- Cozza G, Pinna LA. Casein kinases as potential therapeutic targets. Expert Opin Ther Targets 2015;1-22. [PubMed]
- Battistutta R. Protein kinase CK2 in health and disease:Structural bases of protein kinase CK2 inhibition. Cell Mol Life Sci 2009;66:1868-89. [PubMed]
- Filhol O, Cochet C. Cellular functions of Protein kinase CK2:a dynamic affair. Cell Mol Life Sci 2009;66:1830-9. [PubMed]
- Montenarh M. Protein kinase CK2 and angiogenesis. Advances in Clinical and Experimental Medicine 2014;23:153-8. [PubMed]
- Volodina IuL, Shtil AA. Casein kinase 2, the versatile regulator of cell survival. Mol Biol (Mosk) 2012;46:423-33. [PubMed]
- Guerra B, Issinger OG. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 1999;20:391-408. [PubMed]
- Morales JC, Carpenter PB. Breaking in a new function for casein kinase 2. Sci Aging Knowledge Environ 2004;2004:pe24.
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J 2003;17:349-68. [PubMed]
- Faust M, Montenarh M. Subcellular localization of protein kinase CK2:A key to its function? Cell Tissue Res 2000;301:329-40. [PubMed]
- Welker S, Servas C, Meng R, et al. Tissue specific functions and regulation of protein kinase CK2. In:Ahmed K, Issinger OG, Szyszka R (eds). Protein Kinase CK2 Cellular Function in Normal and Disease States Springer, Cham; Heidelberg; New York; Dordrecht; London, 2015;109-23.
- Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science 2002;298:1912-34. [PubMed]
- Filhol O, Giacosa S, Wallez Y, et al. Protein kinase CK2 in breast cancer:the CK2beta regulatory subunit takes center stage in epithelial plasticity. Cell Mol Life Sci 2015;72:3305-22. [PubMed]
- Vilk G, Weber JE, Turowec JP, et al. Protein kinase CK2 catalyzes tyrosine phosphorylation in mammalian cells. Cell Signal 2008;20:1942-51. [PubMed]
- Wilson LK, Dhillon N, Thorner J, et al. Casein kinase II catalyzes tyrosine phosphorylation of the yeast nucleolar immunophilin Fpr3. J Biol Chem 1997;272:12961-7. [PubMed]
- Basnet H, Su XB, Tan Y, et al. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 2014;516:267-71. [PubMed]
- Christmann M, Tomicic MT, Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutSalpha mismatch-binding activity. Nucleic Acids Res 2002;30:1959-66. [PubMed]
- Utley RT, Lacoste N, Jobin-Robitaille O, et al. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol 2005;25:8179-90. [PubMed]
- Olsen BB, Wang SY, Svenstrup TH, et al. Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage. BMC Mol Biol 2012;13:7. [PubMed]
- Ayoub N, Jeyasekharan AD, Bernal JA, et al. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 2008;453:682-6. [PubMed]
- Guo M, Liu C, Qi FJ, et al. Elevated expression of nuclear protein kinase CK2alpha as a poor prognosis indicator in lymph node cancerous metastases of human thyroid cancers. Asian Pac J Cancer Prev 2014;15:7425-32. [PubMed]
- Zwicker F, Ebert M, Huber PE, et al. A specific inhibitor of protein kinase CK2 delays gamma-H2Ax foci removal and reduces clonogenic survival of irradiated mammalian cells. Radiat Oncol 2011;6:15. [PubMed]
- Lebrin F, Bianchini L, Rabilloud T, et al. CK2α -protein phosphatase 2A molecular complex:Possible interaction with the MAP kinase pathway. Mol Cell Biochem 1999;191:207-12. [PubMed]
- Teitz T, Eli D, Penner M, et al. Expression of the cDNA for the beta subunit of human casein kinase II confers partial UV resistance on xeroderma pigmentosum cells. Mutat Res 1990;236:85-97. [PubMed]
- Kraiss S, Barnekow A, Montenarh M. Protein kinase activity associated with immunopurified p53 protein. Oncogene 1990;5:845-55. [PubMed]
- Meek DW, Simon S, Kikkawa U, et al. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J 1990;9:3253-60. [PubMed]
- Filhol O, Baudier J, Delphin C, et al. Casein kinase II and the tumor suppressor protein P53 associate in a molecular complex that is negatively regulated upon P53 phosphorylation. J Biol Chem 1992;267:20577-83. [PubMed]
- Filhol O, Baudier J, Chambaz EM, et al. Casein kinase 2 inhibits the renaturation of complementary DNA strands mediated by p53 protein. Biochem J 1996;316:331-5. [PubMed]
- Schuster N, Prowald A, Schneider E, et al. Regulation of p53 mediated transactivation by the β-subunit of protein kinase CK2. FEBS Lett 1999;447:160-6. [PubMed]
- Schuster N, Götz C, Faust M, et al. Wild-type p53 inhibits protein kinase CK2 activity. J Cell Biochem 2001;81:172-83. [PubMed]
- Rossi R, Villa A, Negri C, et al. The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J 1999;18:5745-54. [PubMed]
- Herrmann CP, Kraiss S, Montenarh M. Association of casein kinase II with immunopurified p53. Oncogene 1991;6:877-84. [PubMed]
- Lorenz A, Herrmann C, Issinger O, et al. Phosphorylation of wild-type and mutant phenotypes of p53 by an associated protein-kinase. Int J Oncol 1992;1:571-9. [PubMed]
- Kanno S, Kuzuoka H, Sasao S, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J 2007;26:2094-103. [PubMed]
- Shieh SY, Ikeda M, Taya Y, et al. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997;91:325-34. [PubMed]
- Bruins W, Zwart E, Attardi LD, et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 2004;24:8884-94. [PubMed]
- Orphanides G, Wu WH, Lane WS, et al. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 1999;400:284-8. [PubMed]
- Keller DM, Zeng XY, Wang Y, et al. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell 2001;7:283-92. [PubMed]
- Keller DM, Lu H. P53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem 2002;277:50206-13. [PubMed]
- Krohn NM, Stemmer C, Fojan P, et al. Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J Biol Chem 2003;278:12710-5. [PubMed]
- Yamane K, Kinsella TJ. Casein kinase 2 regulates both apoptosis and the cell cycle following DNA damage induced by 6-thioguanine. Clin Cancer Res 2005;11:2355-63. [PubMed]
- Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009-17. [PubMed]
- Morales JC, Franco S, Murphy MM, et al. 53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis. Proc Natl Acad Sci U S A 2006;103:3310-5. [PubMed]
- Morales JC, Xia Z, Lu T, et al. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J Biol Chem 2003;278:14971-7. [PubMed]
- Guerra B, Iwabuchi K, Issinger OG. Protein kinase CK2 is required for the recruitment of 53BP1 to sites of DNA double-strand break induced by radiomimetic drugs. Cancer Lett 2014;345:115-23. [PubMed]
- Yacoub A, Kelley MR, Deutsch WA. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res 1997;57:5457-9. [PubMed]
- Jayaraman L, Murthy KG, Zhu C, et al. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev 1997;11:558-70. [PubMed]
- Fritz G, Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene 1999;18:1033-40. [PubMed]
- Jhanwar-Uniyal M. BRCA1 in cancer, cell cycle and genomic stability. Front Biosci 2003;8:s1107-17. [PubMed]
- Canning MT, Brown DA, Yarosh DB. A bicyclic monoterpene diol and UVB stimulate BRCA1 phosphorylation in human keratinocytes. Photochem Photobiol 2003;77:46-51. [PubMed]
- O'Brien KA, Lemke SJ, Cocke KS, et al. Casein kinase 2 binds to and phosphorylates BRCA1. Biochem Biophys Res Commun 1999;260:658-64. [PubMed]
- Critchfield JW, Coligan JE, Folks TM, et al. Casein kinase II is a selective target of HIV-1 transcriptional inhibitors. Proc Natl Acad Sci USA 1997;94:6110-5. [PubMed]
- Altmannova V, Eckert-Boulet N, Arneric M, et al. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res 2010;38:4708-21. [PubMed]
- Choi BH, Chen Y, Dai W. Chromatin PTEN is involved in DNA damage response partly through regulating Rad52 sumoylation. Cell Cycle 2013;12:3442-7. [PubMed]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993;362:709-15. [PubMed]
- Munshi PN, Lubin M, Bertino JR. 6-thioguanine:a drug with unrealized potential for cancer therapy. Oncologist 2014;19:760-5. [PubMed]
- Parker AR, O'Meally RN, Sahin F, et al. Defective human MutY phosphorylation exists in colorectal cancer cell lines with wild-type MutY alleles. J Biol Chem 2003;278:47937-45. [PubMed]
- Coin F, Auriol J, Tapias A, et al. Phosphorylation of XPB helicase regulates TFIIH nucleotide excision repair activity. EMBO J 2004;23:4835-46. [PubMed]
- Schneider E, Kartarius S, Schuster N, et al. The cyclin H/cdk7/Mat1 kinase activity is regulated by CK2 phosphorylation of cyclin H. Oncogene 2002;21:5031-7. [PubMed]
- Araki M, Masutani C, Takemura M, et al. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem 2001;276:18665-72. [PubMed]
- Trojan P, Rausch S, Giessl A, et al. Light-dependent CK2-mediated phosphorylation of centrins regulates complex formation with visual G-protein. Biochim Biophys Acta 2008;1783:1248-60.
- Grecu D, Assairi L. CK2 phosphorylation of human centrins 1 and 2 regulates their binding to the DNA repair protein XPC, the centrosomal protein Sfi1 and the phototransduction protein transducin beta. FEBS Open Bio 2014;4:407-19. [PubMed]
- Thacker J, Zdzienicka MZ. The XRCC genes:expanding roles in DNA double-strand break repair. DNA Repair (Amst) 2004;3:1081-90. [PubMed]
- London RE. The structural basis of XRCC1-mediated DNA repair. DNA Repair (Amst) 2015;30:90-103. [PubMed]
- Wang W, Lindsey-Boltz LA, Sancar A, et al. Mechanism of stimulation of human DNA ligase I by the Rad9-rad1-Hus1 checkpoint complex. J Biol Chem 2006;281:20865-72. [PubMed]
- Prigent C, Lasko DD, Kodama K, et al. Activation of mammalian DNA ligase through phosphorylation by casein kinase II. EMBO J 1992;11:2925-33. [PubMed]
- Parsons JL, Dianova II, Finch D, et al. XRCC1 phosphorylation by CK2 is required for its stability and efficient DNA repair. DNA Repair (Amst) 2010;9:835-41. [PubMed]
- Kubota Y, Nash RA, Klungland A, et al. Reconstitution of DNA base excision-repair with purified human proteins:interaction between DNA polymerase beta and the XRCC1 protein. EMBO J 1996;15:6662-70. [PubMed]
- Loizou JI, El-Khamisy SF, Zlatanou A, et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 2004;117:17-28. [PubMed]
- Marintchev A, Mullen MA, Maciejewski MW, et al. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat Struct Biol 1999;6:884-93. [PubMed]
- Mani RS, Yu Y, Fang S, et al. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J Biol Chem 2010;285:37619-29. [PubMed]
- Kubota Y, Takanami T, Higashitani A, et al. Localization of X-ray cross complementing gene 1 protein in the nuclear matrix is controlled by casein kinase II-dependent phosphorylation in response to oxidative damage. DNA Repair (Amst) 2009;8:953-60. [PubMed]
- Ström CE, Mortusewicz O, Finch D, et al. CK2 phosphorylation of XRCC1 facilitates dissociation from DNA and single-strand break formation during base excision repair. DNA Repair (Amst) 2011;10:961-9. [PubMed]
- Zhu G, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry 2009;48:4916-25. [PubMed]
- Siddiqui-Jain A, Bliesath J, Macalino D, et al. CK2 inhibitor CX-4945 suppresses DNA repair response triggered by DNA-targeted anticancer drugs and augments efficacy:mechanistic rationale for drug combination therapy. Mol Cancer Ther 2012;11:994-1005. [PubMed]
- Xu W, Chen Q, Wang Q, et al. JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. Cell Death Dis 2014;5:e1551.
- Chen R, Qiu W, Liu Z, et al. Identification of JWA as a novel functional gene responsive to environmental oxidative stress induced by benzo[a]pyrene and hydrogen peroxide. Free Radic Biol Med 2007;42:1704-14. [PubMed]
- Wang S, Gong Z, Chen R, et al. JWA regulates XRCC1 and functions as a novel base excision repair protein in oxidative-stress-induced DNA single-strand breaks. Nucleic Acids Res 2009;37:1936-50. [PubMed]
- Bai J, Zhang J, Wu J, et al. JWA regulates melanoma metastasis by integrin alphaVbeta3 signaling. Oncogene 2010;29:1227-37. [PubMed]
- Wu PY, Frit P, Meesala S, et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 2009;29:3163-3172. [PubMed]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 2009;417:639-50. [PubMed]
- Olsen BB, Issinger OG, Guerra B. Regulation of DNA-dependent protein kinase by protein kinase CK2 in human glioblastoma cells. Oncogene 2010;29:6016-26. [PubMed]
- Olsen BB, Fischer U, Rasmussen TL, et al. Lack of the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is accompanied by increased CK2alpha' levels. Mol Cell Biochem 2011;356:139-47. [PubMed]
- Kim YK, Lee KJ, Jeon H, et al. Protein kinase CK2 is inhibited by human nucleolar phosphoprotein p140 in an inositol hexakisphosphate-dependent manner. J Biol Chem 2006;281:36752-7. [PubMed]
- Solyakov L, Cain K, Tracey BM, et al. Regulation of casein kinase-2 (CK2) activity by inositol phosphates. J Biol Chem 2004;279:43403-10. [PubMed]
- Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol 2015;117:182-93. [PubMed]
- Melander F, Bekker-Jensen S, Falck J, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol 2008;181:213-26. [PubMed]
- Spycher C, Miller ES, Townsend K, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol 2008;181:227-40. [PubMed]
- Jones NC, Farlie PG, Minichiello J, et al. Detection of an appropriate kinase activity in branchial arches I and II that coincides with peak expression of the Treacher Collins syndrome gene product, treacle. Hum Mol Genet 1999;8:2239-45. [PubMed]
- Becherel OJ, Jakob B, Cherry AL, et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res 2010;38:1489-503. [PubMed]
- Dodson GE, Limbo O, Nieto D, et al. Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell Cycle 2010;9:1516-22. [PubMed]
- Yata K, Lloyd J, Maslen S, et al. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol Cell 2012;45:371-83. [PubMed]
- Peng B, Wang J, Hu Y, et al. Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res 2015;43:5936-47. [PubMed]
- Larsen AK, Escargueil AE, Skladanowski A. From DNA damage to G2 arrest: the many roles of topoisomerase II. Prog Cell Cycle Res 2003;5:295-300. [PubMed]
- Wardlaw CP, Carr AM, Oliver AW. TopBP1:A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair (Amst) 2014;22:165-74. [PubMed]
- Rappas M, Oliver AW, Pearl LH. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 2011;39:313-24. [PubMed]
- Takeishi Y, Ohashi E, Ogawa K, et al. Casein kinase 2-dependent phosphorylation of human Rad9 mediates the interaction between human Rad9-Hus1-Rad1 complex and TopBP1. Genes Cells 2010;15:761-71. [PubMed]
- Ohashi E, Takeishi Y, Ueda S, et al. Interaction between Rad9-Hus1-Rad1 and TopBP1 activates ATR-ATRIP and promotes TopBP1 recruitment to sites of UV-damage. DNA Repair (Amst) 2014;21:1-11. [PubMed]
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A 2003;100:1633-8. [PubMed]
- Kroonen J, Artesi M, Capraro V, et al. Casein kinase 2 inhibition modulates the DNA damage response but fails to radiosensitize malignant glioma cells. Int J Oncol 2012;41:776-82. [PubMed]
- Zhao T, Jia H, Li L, et al. Inhibition of CK2 enhances UV-triggered apoptotic cell death in lung cancer cell lines. Oncol Rep 2013;30:377-84. [PubMed]
- Kato T Jr, Delhase M, Hoffmann A, et al. CK2 is a C-terminal IkappaB kinase responsible for NF-kappaB activation during the UV response. Mol Cell 2003;12:829-39. [PubMed]
- Hill R, Lee PW. The DNA-dependent protein kinase (DNA-PK):More than just a case of making ends meet? Cell Cycle 2010;9:3460-69. [PubMed]
- Finka A, Sharma SK, Goloubinoff P. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front Mol Biosci 2015;2:29. [PubMed]
- Kenny MK, Mendez F, Sandigursky M, et al. Heat shock protein 70 binds to human apurinic/apyrimidinic endonuclease and stimulates endonuclease activity at abasic sites. J Biol Chem 2001;276:9532-6. [PubMed]
- Bases R. Heat shock protein 70 enhanced deoxyribonucleic acid base excision repair in human leukemic cells after ionizing radiation. Cell Stress Chaperones 2006;11:240-9. [PubMed]
- Duan Y, Huang S, Yang J, et al. HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2. Cell Stress Chaperones 2014;19:271-9. [PubMed]
- Komander D, Clague MJ, Urbe S. Breaking the chains:structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 2009;10:550-63. [PubMed]
- Nakada S, Tai I, Panier S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 2010;466:941-6. [PubMed]
- Herhaus L, Perez-Oliva AB, Cozza G, et al. Casein kinase 2 (CK2) phosphorylates the deubiquitylase OTUB1 at Ser16 to trigger its nuclear localization. Sci Signal 2015;8:ra35. [PubMed]


