
Mechanism of periplakin on ovarian cancer cell phenotype and its influence on prognosis
Introduction
Ovarian cancer (OV) is the second most common malignant tumor in women after breast cancer, the seventh most common female cancer in the world, and the fifth leading cause of cancer-related deaths in women, with 21,410 new cases and 13,770 deaths projected to occur in the United States in 2021 (1). The prognosis of OV is directly related to the stage of the disease at the time of diagnosis. Most women are diagnosed with advanced disease and relapse after radical surgery and chemotherapy. For patients with stage I OV, the 5-year survival rate is 90%, which decreases to about 80% in patients with local disease and 25% in patients with metastatic disease (1). The current clinical treatment strategy for OV includes surgical resection and systemic treatment. Although some progress has been made, treatment strategies are still limited, and the prognosis of OV is very poor(2). The etiology and pathogenesis of OV remain unclear, although epidemiological investigation has shown that hormones, reproduction, heredity factors, inflammation, living habits, and other factors can affect the occurrence of OV(3,4). A study has shown that OV is a heterogeneous disease with complex molecular and gene changes(5). Therefore, studying the key molecular changes in its pathogenesis is of great clinical significance.
The plakin protein family is a family of 7 cytoskeletal proteins [microtubule-actin crosslinking factor 1 (MACF1), bullous pemphigoid antigen 1 (BPAG1), agglutinin, desmoplakin, envoplakin, periplakin (PPL), and epiplakin](6), which are located in desmosomes and intermediate filaments and form a cellular “molecular bridge” between the membrane-intracellular cytoskeleton and the cell-cell junction(7). The plakin protein family is closely related to the development of malignant tumors. Its family members are involved in many biological processes that regulate tumor proliferation, invasion, metastasis, and resistance. Therefore, these proteins can be used as drug targets for tumor prognosis and antitumor therapy (8-10).
One of these family members, PPL, is a 195 kDa membrane-related protein mainly located in the desmosomes and desmosome interstitial membranes of differentiated epidermal keratinocytes. PPL is highly homologous to other members of the plakin family. Its function is to initiate the formation of the keratinized capsule, a layer of crosslinked protein formed under the plasma membrane during the differentiation of keratinocytes that forms the permeability barrier of the skin. PPL is widely expressed in keratinized and nonkeratinized epithelial cells, such as skin, mouth, and bladder cells (11), and is associated with the onset of a fatal autoimmune blister disease associated with malignant tumors (12). Although its role in tumors has not been well studied, PPL has been reported to play a tumor suppressive role in a variety of tumors, such as urothelial tumors and colorectal cancer. One study found that PPL protein was highly expressed in urinary epithelial tumor tissue (11). Another study found that PPL expression was significantly decreased in colon cancer compared with that in adjacent tissues, and that its low expression was related to tumor size. This study showed that PPL induced cell cycle arrest to inhibit the proliferation ability of colorectal cancer cells, inhibited tumor cell epithelial-mesenchymal transition, and reduced invasion and metastasis (13). Moreover, PPL binds to protein kinase B (AKT), acting as a localization signal in the AKT signaling pathway (14).
At present, PPL expression in broad-spectrum tumors and its influence on patient survival are not well understood, especially with respect to OV. Therefore, this paper aimed to explore the influence of PPL expression on OV and its molecular mechanism. PPL expression in various tumors and its relationship with prognosis were analyzed using The Cancer Genome Atlas (TCGA), the Gene Expression Omnibus (GEO), and the DepMap database, and the role of PPL in OV was studied to provide an experimental basis for the use of PPL as a candidate molecular target for treatment. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1090/rc).
Methods
Bioinformatics analysis
A total of 31 tumor types and their matched normal tissue samples containing epigenomic, transcriptomic, and proteomic data were extracted from TCGA (http://ualcan.path.uab.edu/index.html). Gene expression profiling data were extracted from GEO (https://www.ncbi.nlm.nih.gov/). The Depmap database (https://depmap.org/portal/) was a database consisting of cell line CRISPR screening data that could be used to analyze the effect of genes on the ability of tumor cell lines to grow using the Depmap database.
Clinical samples
OV patients treated in the Department of Gynecology at The Third Affiliated Hospital of Qiqihar Medical College from January 2019 to December 2020 were included in the clinical samples. The inclusion criteria were as follows: (I) patients with ovary cancer first surgical resection; (II) no other anti-tumor treatment was performed before the operation; and (III) complete follow-up data were included in the pathological data. Clinicopathological characteristics of these patients are shown in Table S1. Three pairs of OV and matched adjacent normal tissues were obtained from the operating room. After phosphate buffered saline (PBS) washing, each tissue was frozen in liquid nitrogen and combined for subsequent experimental assays. Clinicopathological characteristics of the 3 patients are shown in Table S2. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of The Third Affiliated Hospital of Qiqihar Medical University (No. 2021LL-2), and informed consent was taken from all the patients.
Cell culture
OV cell lines, including OVCAR4, CAOV-4, CoC1, OVMANA, EFO21, and EFO27, were cultured for resuscitation and centrifuged at 1,000 rpm for 5 minutes. The cells were resuspended in Dulbecco’s Modified Eagle Medium (DMEM; HyClone, Cytiva, Marlborough, MA, USA) containing 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and placed in a cell culture flask for incubation at 37 ℃ with 5% CO2 in a cell incubator. Every 48 hours, the complete medium was replaced. When the cell culture reached 80% confluence, the cells were subcultured according to a 1:2 ratio and then stably transmitted for 3–4 generations for subsequent cell experiments.
RT-qPCR
When the cells were cultured to 80%, they were washed with PBS 3 times, and TRIzol reagent (Invitrogen, Waltham, MA USA) was added to lyse the cells. The total RNA was extracted from the cells with chloroform, isopropanol, and alcohol and reverse transcribed to complementary DNA (cDNA) with a SYBR Premix Ex TaqTM quantitative reverse transcription-polymerase chain reaction (RT-qPCR) kit (Takara Bio Inc., Shiga, Japan). According to the manufacturer’s instructions, RT-qPCR was performed at 95.94 ℃ for 15 seconds, at 60 ℃ for 20 seconds, and at 72 ℃ for 10 seconds, for 40 cycles. The threshold cycle (Ct) for each sample was detected. Using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the internal control, the relative expression of PPL messenger RNA (mRNA) was calculated using the delta-delta Ct (2–∆∆Ct) method. The primers are shown in Table 1.
Table 1
| Primer name | Primer sequence |
|---|---|
| PPL | 5'- AGGCAAATACAGCCCCACTG -3' (Forward) |
| 5'- AGGTCACTCTGCATCTTGGC -3' (Reverse) | |
| GAPDH | 5'- GTGAACCATGAGAAGTATG -3' (Forward) |
| 5'- CGGCCATCACGCCACAGTTTC -3' (Reverse) |
PPL, periplakin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
PPL small interfering RNA (siRNA) transient transfection
Transient transfection siRNA oligonucleotide PPL (si-PPL) and negative controls (si-NC) were purchased from Thermo Fisher Scientific. The cells were cultured to 30–40% confluence and divided into si-NC, si-PPL-1, si-PPL-2 and si-PPL-3 groups. Each group was transfected with 20 pmol of siRNA oligonucleotides using Lipofectamine 2000 reagents (Invitrogen, Carlsbad, CA, USA). After transfection for 48 hours, the RNA of the cells was extracted, and PPL expression in each group, as well as the inhibition of siRNA on PPL, was detected with RT-qPCR. Two siRNA sequences with obvious inhibitory effects were selected for subsequent studies.
MTT detection of cell proliferation
When OV cells were overgrown with the highest and the lowest expression of PPL, cells were counted after trypsin digestion, inoculated into a 96-well plate at 3,000 cells/well with 6 parallel holes in each group, and cultured with 5% CO2 at 37 ℃ in an incubator. siRNA was transfected according to the above transfection method. After 48 hours of transfection, 100 µL of fresh media was replaced, with 20 µL MTT reagent added (Beyotime Biotechnology, Shanghai, China). After another 4 hours of culture, the culture media was sucked and discarded, and 150 µL dimethyl sulfoxide (DMSO) was added to dissolve for crystallization. The optical density (OD) value of each hole at 490 nm was detected using a microplate reader. The cell proliferation rate was calculated according to the following formula: (1 − OD value of the experimental group/the OD value of the control group) × 100%.
Flow cytometry detection of apoptosis
Cells and floating cells in cell medium after transfection with siRNA for 48 hours were collected from each group with a cell scraper, centrifuged at 2,000 rpm for 5 minutes, and washed with PBS 3 times. A 500 µL staining buffer was added to each well to resuspend the cells, 5 µL Annexin V-FITC and 5 µL PI staining reagents (Beyotime Biotechnology) were added for apoptosis staining, and the cells were incubated at room temperature for 15 minutes. The apoptosis rate of each group was detected by flow cytometry.
Western blotting
Cells were collected from each group after transfection with siRNA for 48 hours, centrifuged at 2,000 rpm for 5 minutes, and washed with PBS 3 times, with protein lysis reagents added. After 20 minutes of lysis on ice, the cells were centrifuged at 14,000 rpm and 4 ℃ for 30 minutes. Cell debris was removed, and the protein concentration was detected. The protein was denatured by boiling for 3 minutes, and the target protein was separated by gel electrophoresis. Next, the protein was wet transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, MilliporeSigma, Burlington, MA, USA) and incubated with 10% skimmed milk at room temperature for 1 hour. Primary antibodies were incubated at 4 ℃ overnight, and secondary antibodies were incubated at room temperature for 1 hour. The protein band was exposed using an ECL kit (Thermo Fisher Scientific). Name, cargo number, and the working concentration of antibodies used in the experiment are shown in Table 2.
Table 2
| Name of antibody | Company | Cargo number | Working concentration |
|---|---|---|---|
| PPL | Abcam, UK | ab131269 | 1:1,000 |
| Cleaved Caspase-3 | Abcam, UK | ab2302 | 1:500 |
| BAX | Abcam, UK | ab32503 | 1:5,000 |
| BCL-2 | Abcam, UK | ab32124 | 1:1,000 |
| GAPDH | Abcam, UK | ab181602 | 1:1,000 |
| Goat Anti-Mouse IgG | Bioss, Beijing | bs-0296G | 1:3,000 |
| Goat Anti-rabbit IgG | Bioss, Beijing | bs-0295G | 1:3,000 |
PPL, periplakin; BAX, BCL-2-associated X; BCL-2, B-cell lymphoma 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Statistical analysis
SPSS 17.0 statistics software (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The difference in the PPL positive expression rate between OV and normal ovarian tissue and the relationship between PPL and the clinicopathological parameters of OV patients were analyzed by chi-square test. The influence of PPL on the prognosis of OV patients was analyzed by logrank test. Differences between 2 groups were analyzed using a t test, and differences among 3 groups were analyzed by analysis of variance (ANOVA). P<0.05 was considered statistically significant.
Results
PPL expression in tumor tissues
PPL expression in 31 malignant tumor tissues and its relationship with patient prognosis were analyzed in TCGA database using bioinformatics methods. The results suggested that PPL expression might play an important role in the occurrence and development of malignant tumors. Compared with the control group, PPL mRNA expression was significantly upregulated in cholangiocarcinoma (CHOL), OV, pancreatic adenocarcinoma (PAAD), and thymic carcinoma (THYM) tumors (Figure 1A). PPL mRNA expression was significantly downregulated in breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), low-grade glioma (LGG), kidney chromophobe (KICH), and skin cutaneous melanoma (SKCM) tumors (Supplementary Figure S1A). High expression of PPL was associated with poor survival in LGG and OV but with good survival in adenoid cystic carcinoma (ACC) and sarcoma (SARC; Supplementary Figure S1B). These results suggested that high PPL expression in OV is associated with poor prognosis (Figure 1B), and that PPL might therefore play an important role in OV.
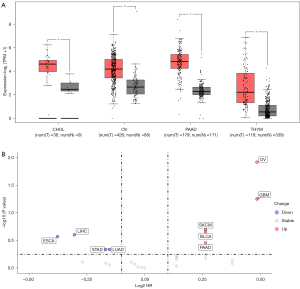
PPL expression is high in OV and is associated with prognosis
To further validate the results of the TCGA analysis, the data from the GEO database were analyzed for PPL expression. The results of the GSE6008 data analysis showed that compared with 4 control groups, PPL was highly expressed in 99 cases of OV tissues, with a Log2 (FC) of 0.51 (P=0.0042; Figure 2A). Moreover, Kaplan-Meier survival analysis of OV samples from TCGA showed that overall survival (OS; P=0.014) and progress-free survival (PFS; P=0.0033) of patients in the high expression PPL group were worse than those of patients in the low expression PPL group (Figure 2B,2C).
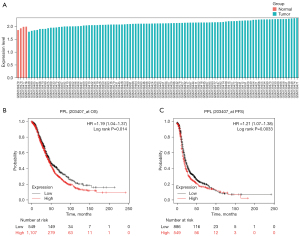
Relationship between PPL expression and prognosis in clinical OV tissues
PPL expression in 70 cases of OV tissues and 30 cases of normal ovarian tissues was detected by western blotting. The results showed that PPL expression in OV tissues was higher than that in normal ovarian tissues (P<0.05; Figure 3A,3B). The relationship between PPL expression and clinicopathological characteristics of patients with OV was analyzed using a chi-square test. The results showed that high PPL expression was associated with histological grading (P=0.016), International Federation of Gynecology and Obstetrics (FIGO) staging (P=0.018), and lymph node metastasis (P=0.011) but was independent of patient age and tissue type (P>0.05; Table 3). To analyze the influence of PPL expression on the prognosis of OV patients, a survival curve of the patients was drawn using SPSS software, and the results showed that patients with high PPL expression had a worse prognosis than those with low PPL expression (P=0.011; Figure 3C). Univariate regression analysis showed a statistically significant relationship between PPL expression and survival and lymph node metastasis and survival (Table S3). Finally, multivariate regression analysis showed that the combination of PPL expression and lymph node metastasis did not reach significance when predicting OV survival (Table S3).
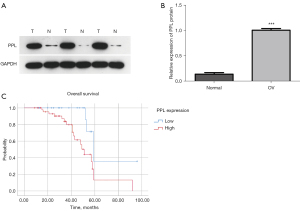
Table 3
| Characteristics | PPL expression | P value | |
|---|---|---|---|
| Low, no. cases | High, no. cases | ||
| Age (years) | 0.552 | ||
| ≤50 | 12 | 36 | |
| >50 | 7 | 15 | |
| Histopathological type | 0.596 | ||
| Serous carcinoma | 15 | 43 | |
| Mucous carcinoma | 4 | 8 | |
| FIGO stage | 0.018 | ||
| I + II | 8 | 37 | |
| III + IV | 12 | 13 | |
| Histologic grade | 0.016 | ||
| Moderate and well | 7 | 35 | |
| Poor | 12 | 16 | |
| Lymphovascular invasion | 0.011 | ||
| No | 15 | 23 | |
| Yes | 4 | 28 | |
PPL, periplakin; FIGO, International Federation of Gynecology and Obstetrics.
Biological functions of PPL and KEGG pathway analysis in OV cell lines
The influence of PPL on the growth ability of OV cell lines was analyzed using the DepMap database. Based on the results of the CRISPR analysis, PPL genes in the cell line were compared with all the genes in the OV cell lines. The results showed that the PPL gene dependency score was lower, indicating poorer cell growth ability (Figure 4A). In addition, the results of the CRISPR and RNA interference (RNAi) analysis showed that the dependency scores of PPL genes in the 2 common cell lines JHOS2 and SUN8 were negative, and that cell survival was inhibited (Figure 4B). The samples were divided into upregulated and downregulated groups according to the median mRNA expression of PPL in 307 OV samples for Bayesian analysis. Bayesian t values of each gene were obtained by Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway analysis, which showed that PPL was associated with the apoptosis pathway (hsa04210; P<0.001; Table 4).
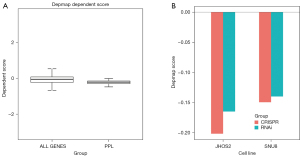
Table 4
| Geneset | Description | Size | Overlap | Expect | ERatio | P value | FDR |
|---|---|---|---|---|---|---|---|
| Hsa04520 | Adherens junction | 72 | 17 | 1.72 | 9.91 | 5.20E-13 | 1.44E-10 |
| Hsa04670 | Leukocyte transendothelial migration | 112 | 14 | 2.67 | 5.25 | 3.58E-07 | 4.96E-05 |
| Hsa04310 | Wnt signaling pathway | 146 | 14 | 3.48 | 4.02 | 8.87E-06 | 7.78E-04 |
| Hsa05226 | Gastric cancer | 149 | 14 | 3.55 | 3.94 | 1.12E-05 | 7.78E-04 |
| Hsa05215 | Prostate cancer | 97 | 11 | 2.31 | 4.76 | 1.74E-05 | 9.61E-04 |
| Hsa04512 | ECM-receptor interaction | 82 | 10 | 1.95 | 5.12 | 2.22E-05 | 1.03E-03 |
| Hsa05213 | Endometrial cancer | 58 | 8 | 1.38 | 5.79 | 6.16E-05 | 1.93E-03 |
| Hsa05222 | Small cell lung cancer | 93 | 10 | 2.22 | 4.51 | 6.70E-05 | 193E-03 |
| Hsa04068 | FoxO signaling pathway | 132 | 12 | 3.15 | 3.81 | 6.71E-05 | 193E-03 |
| Hsa00310 | Lysine degradation | 59 | 8 | 1.41 | 5.69 | 6.98E-05 | 1.93E-03 |
| Hsa04210 | Apoptosis | 136 | 12 | 3.24 | 3.70 | 8.91E-05 | 2.26E-03 |
| Hsa04330 | Notch signaling pathway | 48 | 7 | 1.16 | 6.12 | 1.25E-04 | 2.89E-03 |
| Hsa05224 | Breast cancer | 147 | 12 | 3.50 | 3.43 | 1.89E-04 | 4.03E-03 |
| Hsa05218 | Melanoma | 72 | 8 | 1.72 | 4.66 | 2.90E-04 | 5.73E-03 |
| Hsa05100 | Bacterial invasion of epithelial cells | 74 | 8 | 1.76 | 4.54 | 3.50E-04 | 6.47E-03 |
| Hsa05220 | Chronic myeloid leukemia | 76 | 8 | 1.81 | 4.42 | 4.21E-04 | 6.80E-03 |
| Hsa04919 | Thyroid hormone signaling pathway | 116 | 10 | 2.76 | 3.62 | 4.22E-04 | 6.80E-03 |
| Hsa05146 | Amoeba | 96 | 9 | 2.29 | 3.93 | 4.42E-04 | 6.80E-03 |
| Hsa01521 | EGFR tyrosine kinase inhibitor resistance | 79 | 8 | 1.88 | 4.25 | 5.48E-04 | 7.90E-03 |
| Hsa04916 | Melanogenesis | 101 | 9 | 2.41 | 3.74 | 6.43E-04 | 8.91E-03 |
| Hsa05217 | Basal cell carcinoma | 63 | 7 | 1.50 | 4.66 | 6.97E-04 | 9.19E-03 |
TCGA, The Cancer Genome Atlas; PPL, periplakin; ERatio, Enrichment Ratio; FDR, false discovery rate.
Influence of PPL on proliferation and apoptosis of OV cells
To assess PPL expression in different OV cell lines, we detected PPL expression in 6 OV cell lines (OVCAR4, CAOV-4, CoC1, OVMANA, EFO27, and EFO21). The results showed that PPL mRNA expression was highest in CAOV-4 cells and lowest in CoC1 cells (Figure 5A). We selected CAOV-4 and CoC1 cells for PPL siRNA transfection for subsequent experimental studies and used the PPL siRNA–transfected CAOV-4 and CoC1 cells to build a PPL knockdown model. After transfection for 48 hours, the efficiency of PPL siRNA transfection into the OV cells was verified by RT-qPCR (Figure 5B). Compared with that in the si-NC group, PPL mRNA expression in the si-PPL-1, si-PPL-2, and si-PPL-3 groups of CAOV-4 and CoC1 cells was significantly decreased (P<0.05). The inhibiting influence of the si-PPL-1 and si-PPL-3 group of CAOV-4 and CoC1 cells was significant, and these cells were selected for subsequent functional experiments. To investigate whether PPL had an influence on the proliferation and apoptosis of OV cells, we detected the influence of PPL siRNA on the proliferation of OV cells with MTT. Compared with that of the si-NC group, the proliferation rate of the si-PPL-1 and si-PPL-3 groups of CAOV-4 and CoC1 cells was decreased (P<0.05; Figure 5C). The influence of PPL siRNA on the apoptosis of OV cells was detected by flow cytometry. Compared with that of the si-NC group, the apoptosis rate of the si-PPL-1 and si-PPL-3 groups of CAOV-4 and CoC1 cells was increased (P<0.05; Figure 5D).
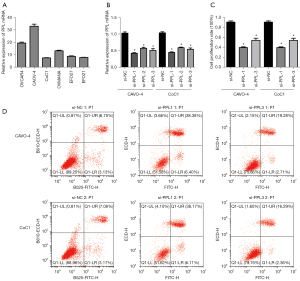
Western blotting of apoptosis-related protein expression
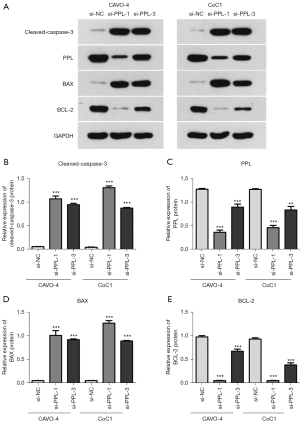
The influence of PPL siRNA on apoptosis-related proteins in OV cells was detected by western blotting. Compared with that in the si-NC group, the expression of active caspase 3 and BCL-2-associated X (BAX) proteins in the si-PPL-1 and si-PPL-3 groups of CAOV-4 and CoC1 cells was increased, while the expression of B-cell lymphoma 2 (BCL-2) protein was decreased (Figure 6). This indicated that knockdown of PPL could induce increased expression of the pro-apoptotic proteins caspase 3 and BAX and inhibit the expression of the anti-apoptotic gene BCL-2, thus promoting the apoptosis of OV cells.
Discussion
OV is a common gynecological tumor with a high degree of malignancy that primarily affects menopausal women (1). Traditional treatment methods, including surgery, radiotherapy, and chemotherapy, can improve the survival of patients with early OV. However, due to the latent clinical symptoms of OV, 70% of patients have stage III or IV disease with extensive metastasis at the time of initial diagnosis. Therefore, late diagnosis is one of the major factors in the high mortality of patients, and advanced and recurrent OV remain the most lethal form of gynecologic malignancy (15). Unlike other tumors, advanced OV tends to invade and metastasize in the peritoneum, with a high degree of malignancy. Traditional treatment methods are limited for patients with advanced OV, and more than half of early OV patients experience recurrence within 16 months of treatment, resulting in a 5-year OS rate of less than 50 percent (2). New molecular targeted therapies have shown promise as a treatment for the clinical symptoms of late and recurrent OV, but the long-term prognosis of patients has not been significantly improved (16). As gene change is a key factor in tumor pathogenesis, studying the pathogenesis of OV at the genetic level is an effective way to find new targets for OV treatment.
PPL gene-encoded proteins are part of the epidermal keratinization envelope of desmosomes and keratinocytes. These proteins provide strong support for terminal differentiated keratinocytes, act as epidermal barriers, and help reorganize keratin intermediate filament networks to facilitate wound closure (17,18). Desmoplakin and PPL have been reported to promote the development of eosinophilic esophagitis by affecting the barrier integrity, cell motility, and Rho GTPase activity of esophageal epithelial cells through mutations (19). Besnard et al. found that PPL attenuated bleomycin-induced lung injury in mice while promoting the progression of lung remodeling (20). PPL has been reported to inhibit tumor malignant progression in urothelial tumors and colorectal cancer tissues (11,13). In addition, serum PPL is a potential biomarker of urinary urothelial carcinoma, and its expression is significantly correlated with the pathological stage and cancer-specific survival of patients (21,22). However, there are still few studies on the role of PPL in tumors, and its biological function is not yet clear. This study used public data from TCGA and GEO to conduct a bioinformatic analysis of the correlation between genes and disease (23,24). PPL expression in 31 malignant tumors and the relationship between PPL expression and patient prognosis were analyzed using TCGA. The results showed that PPL expression was increased in CHOL, OV, PAAD, and THYM tumors and decreased in BRCA, ESCA, GBM, HNSC, LGG, KICH, and SKCM tumors. High PPL expression was associated with poor survival in patients with LGG and OV, and low PPL expression was associated with poor survival in patients with ACC and SARC, indicating that PPL might play different biological functions in different tumors. Highly expressed PPL in OV tissues was associated with poor prognosis in patients with OV. The results of the GEO data analysis showed that PPL expression in OV tissues was significantly higher than that in normal control tissues, and that poor prognosis was associated with high PPL expression. These results suggested that PPL might play an oncogenic role in OV.
We further verified the biological function of PPL at the tissue and cell levels. The expression level of PPL protein in OV tissue and normal ovarian tissue was detected by western blotting. The results showed that PPL protein expression was increased in OV tissues, which was consistent with the results of the database analysis. Chi-square test analysis found that the positive expression rate of PPL was related to OV histological grading, FIGO staging, and lymph node metastasis, suggesting that PPL might be involved in the malignant progression of the disease. The follow-up data of 70 patients with OV were analyzed and a survival curve was drawn. The results showed that the prognosis of patients with high PPL expression was poor compared with that of patients with low PPL expression, which was consistent with the results of the bioinformatics analysis. Univariate regression analysis suggested a significant relationship between PPL expression and lymph node metastasis and survival, while multivariate regression analysis showed that PPL expression did not reach significance when combined with lymph node metastasis to predict OV survival. These results indicated that there might be a complex relationship between PPL expression and lymph node metastasis, but this relationship could not be fully predicted due to the small sample size in our study.
KEGG analysis of PPL and related genes suggested that PPL might be related to the apoptosis pathway of the tumor. Apoptosis is a type of programmed cell death that plays a key role in the normal development of cell organisms and helps to maintain the homeostasis of the body (25). Decreased apoptosis is an important feature of tumors, and the induction of apoptosis is the mechanism of many anti-tumor drugs (26,27). To test whether PPL regulates OV cell apoptosis, we first examined PPL expression in 6 OV cell lines. OV cells with high and low PPL expression were selected for PPL siRNA transfection to construct a knockdown cell model. According to the MTT test of cell proliferation, the proliferation rate of OV cells decreased significantly after PPL expression was inhibited. To further verify whether PPL affects OV apoptosis, we used flow cytometry to detect the apoptosis of OV cells after PPL knockdown and found that silencing PPL increased OV apoptosis. These results suggested that inhibition of PPL expression could promote apoptosis of OV cells. However, the molecular mechanism of its function needs to be further explored.
Caspase 3 protein is the initiator of the caspase 3 apoptotic signaling pathway. When external stimulated, caspase 3 are activated into active caspase 3, which can cascade after activation to promote the activation of downstream apoptotic proteins and the occurrence of apoptosis (28). BCL-2 and BAX belong to the family of BCL-2 apoptotic proteins. BCL-2 is an important anti-apoptotic gene, and BAX promotes apoptosis by inhibiting BCL-2 activity (29,30). Western blotting detection showed that the expression of active caspase 3 and BAX protein in transfected OV cells was increased, while the expression of BCL-2 proteins was decreased, suggesting that PPL might inhibit apoptosis by regulating the expression of the apoptosis-related proteins caspase 3, BAX, and BCL-2.
Conclusions
To sum up, the high expression of PPL in OV tissues was related to the poor prognosis of OV, and PPL might inhibit apoptosis by regulating the expression of apoptosis-related proteins. Therefore, PPL might play a cancer-promoting role in OV and could be a potential molecular target to inhibit malignant progression.
Acknowledgments
Funding: This study was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1090/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1090/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1090/coif). All authors report that this study was funded by the Clinical Research Fund Project of Qiqihar Academy of Medical Sciences (No. QMSI2020L-15). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of The Third Affiliated Hospital of Qiqihar Medical University (No. 2021LL-2), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Stewart C, Ralyea C, Lockwood S. Ovarian Cancer: An Integrated Review. Semin Oncol Nurs 2019;35:151-6. [Crossref] [PubMed]
- Li S, Jiang K, Li J, et al. Estrogen enhances the proliferation and migration of ovarian cancer cells by activating transient receptor potential channel C3. J Ovarian Res 2020;13:20. [Crossref] [PubMed]
- Moufarrij S, Dandapani M, Arthofer E, et al. Epigenetic therapy for ovarian cancer: promise and progress. Clin Epigenetics 2019;11:7. [Crossref] [PubMed]
- Grunewald T, Ledermann JA. Targeted Therapies for Ovarian Cancer. Best Pract Res Clin Obstet Gynaecol 2017;41:139-52. [Crossref] [PubMed]
- Ruhrberg C, Hajibagheri MA, Parry DA, et al. Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J Cell Biol 1997;139:1835-49. [Crossref] [PubMed]
- Odintsova E, Mohammed F, Trieber C, et al. Binding of the periplakin linker requires vimentin acidic residues D176 and E187. Commun Biol 2020;3:83. [Crossref] [PubMed]
- Quick QA. Microtubule-Actin Crosslinking Factor 1 and Plakins as Therapeutic Drug Targets. Int J Mol Sci 2018;19:368. [Crossref] [PubMed]
- Wesley T, Berzins S, Kannourakis G, et al. The attributes of plakins in cancer and disease: perspectives on ovarian cancer progression, chemoresistance and recurrence. Cell Commun Signal 2021;19:55. [Crossref] [PubMed]
- Hu L, Huang Z, Wu Z, et al. Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer. Int J Mol Sci 2018;19:974. [Crossref] [PubMed]
- Hasegawa Y, Kamata Y, Yorozu T, et al. Expression of plakin family in urothelial carcinoma of the upper urinary tract. Nihon Hinyokika Gakkai Zasshi 2017;108:87-95. [Crossref] [PubMed]
- Maruta CW, Miyamoto D, Aoki V, et al. Paraneoplastic pemphigus: a clinical, laboratorial, and therapeutic overview. An Bras Dermatol 2019;94:388-98. [Crossref]
- Li X, Zhang G, Wang Y, et al. Loss of periplakin expression is associated with the tumorigenesis of colorectal carcinoma. Biomed Pharmacother 2017;87:366-74. [Crossref] [PubMed]
- van den Heuvel AP, de Vries-Smits AM, van Weeren PC, et al. Binding of protein kinase B to the plakin family member periplakin. J Cell Sci 2002;115:3957-66. [Crossref] [PubMed]
- Bookman MA. Optimal primary therapy of ovarian cancer. Ann Oncol 2016;27:i58-62. [Crossref] [PubMed]
- Guan LY, Lu Y. New developments in molecular targeted therapy of ovarian cancer. Discov Med 2018;26:219-29. [PubMed]
- Long HA, Boczonadi V, McInroy L, et al. Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J Cell Sci 2006;119:5147-59. [Crossref] [PubMed]
- Gujrati M, Mittal R, Ekal L, et al. SUMOylation of periplakin is critical for efficient reorganization of keratin filament network. Mol Biol Cell 2019;30:357-69. [Crossref] [PubMed]
- Shoda T, Kaufman KM, Wen T, et al. Desmoplakin and periplakin genetically and functionally contribute to eosinophilic esophagitis. Nat Commun 2021;12:6795. [Crossref] [PubMed]
- Besnard V, Dagher R, Madjer T, et al. Identification of periplakin as a major regulator of lung injury and repair in mice. JCI Insight 2018;3:90163. [Crossref] [PubMed]
- Matsumoto K, Ikeda M, Matsumoto T, et al. Serum periplakin as a potential biomarker for urothelial carcinoma of the urinary bladder. Asian Pac J Cancer Prev 2014;15:9927-31. [Crossref] [PubMed]
- Matsumoto K, Ikeda M, Sato Y, et al. Loss of periplakin expression is associated with pathological stage and cancer-specific survival in patients with urothelial carcinoma of the urinary bladder. Biomed Res 2014;35:201-6. [Crossref] [PubMed]
- Ye B, Shi J, Kang H, et al. Advancing Pan-cancer Gene Expression Survial Analysis by Inclusion of Non-coding RNA. RNA Biol 2020;17:1666-73. [Crossref] [PubMed]
- Wang S, Yin C, Zhang Y, et al. Overexpression of ICAM-1 Predicts Poor Survival in High-Grade Serous Ovarian Carcinoma: A Study Based on TCGA and GEO Databases and Tissue Microarray. Biomed Res Int 2019;2019:2867372. [Crossref] [PubMed]
- Dupont-Versteegden EE. Apoptosis in skeletal muscle and its relevance to atrophy. World J Gastroenterol 2006;12:7463-6. [Crossref] [PubMed]
- Lin J, Song T, Li C, et al. GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim Biophys Acta Mol Cell Res 2020;1867:118659. [Crossref] [PubMed]
- Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol 2020;17:395-417. [Crossref] [PubMed]
- Boice A, Bouchier-Hayes L. Targeting apoptotic caspases in cancer. Biochim Biophys Acta Mol Cell Res 2020;1867:118688. [Crossref] [PubMed]
- Tambaro FP, Wierda WG. Tumour lysis syndrome in patients with chronic lymphocytic leukaemia treated with BCL-2 inhibitors: risk factors, prophylaxis, and treatment recommendations. Lancet Haematol 2020;7:e168-76. [Crossref] [PubMed]
- Walensky LD. Targeting BAX to drug death directly. Nat Chem Biol 2019;15:657-65. [Crossref] [PubMed]
(English Language Editor: C. Gourlay)


