
Histone methyltransferase KMT2D mediated lipid metabolism via peroxisome proliferator-activated receptor gamma in prostate cancer
Introduction
Currently, prostate cancer (PCa) is the most common type of cancer in men, with more than 350,000 related deaths worldwide every year (1). It has become one of the hot spots in the field of cancer research in men. There are various treatment methods for early localized PCa, which include radical surgical resection, radiotherapy, chemotherapy, and androgen deprivation therapy (ADT) (2). Life expectancy may be as high as 99% over 10 years for men with localized PCa, as well as in patients who are actively receiving treatment, if diagnosed at an early stage (1). However, patients with advanced PCa (distant metastases) have a poor overall survival rate of only 30% over 5 years (1). There is an urgent need to further explore PCa treatment direction. A recent study reported that the levels of more than 20 lipid metabolites are upregulated during PCa development (3). The role of lipid metabolism in PCa development may be more important than expected, and hence, it may be crucial in PCa diagnosis and treatment. An increase in the levels of various lipid metabolizing enzymes, such as fatty acid synthase (FASN), promotes the upregulation of fatty acids, cholesterol, phospholipids, and androgens (4). Additionally, it enhances the invasion ability of cancer cells (5). Although numerous studies have analyzed the role of lipid metabolism in PCa, the specific underlying mechanism remains unclear and requires further exploration (3,6,7).
Histone-lysine N-methyltransferase 2D (KMT2D) is a member of lysine methyltransferase 2, which belongs to the mixed-lineage leukemia family. It methylates histone H3 lysine 4 (H3K4) to promote genome-wide accessibility and transcription (8). Moreover, KMT2D also acts as a major enhancer regulator and translates various biological processes such as regulation of development, differentiation, metabolism, and tumor suppression in mammalian cells (9). Exome sequencing studies have shown that KMT2D is one of the most frequently mutated genes in many types of human cancers (10-14). Our laboratory has focused on KMT2D involvement in PCa pathogenesis (15). In a previous study, it was found that KMT2D is highly expressed in PCa cases and is associated with a poor prognosis. Interestingly, in recent studies of hepatic steatosis, KMT2D plays a key role in this lipid metabolism process and that it may be mobilized to the sequences of multiple target genes related to lipid metabolism by peroxisome proliferator-activated receptor gamma (PPARγ) (16). Adipogenesis and hepatic steatosis share a similar gene regulatory program (17). However, the role and mechanism of KMT2D in lipid metabolism remain unclear in PCa.
Here, KMT2D was found to be directly involved in the regulation of PPARγ lipid metabolism, which affected PCa cell growth and proliferation. Mechanistically, KMT2D promoted oncogenic and abnormal lipid metabolism in PCa cell lines by forming a complex with PPARγ. Thus, the inhibition of KMT2D and PPARγ expression may be an interesting PCa treatment strategy. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-431/rc).
Methods
Cell culture
The Human PCa cell lines of DU145 (CC1201, Cellcook, Guangzhou, China), PC3 (CC1202, Cellcook, Guangzhou, China), and LNCaP (CC1204, Cellcook, Guangzhou, China) were selected for this study. The cells were cultured in RPMI1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD, USA), and were grown in a humidified 37 ℃ atmosphere containing 5% carbon dioxide. All cell lines tested negative for mycoplasma contamination. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reagents
Rosiglitazone (R2408) (ROSI, A PPARγ pathway agonist) and DMSO (D2650) were purchased from Sigma (St. Louis, MO, USA). For culture experiments, 10 µM ROSI was dissolved in DMSO, which was further diluted in RPMI1640 medium and added to the culture to a final DMSO concentration of 0.1%.
KMT2D knockdown by siRNA
As described previously (15), siRNA targeting KMT2D was chemically synthesized (Dharmacon). PC-3, DU-145, and LNCaP cells were transfected with siRNA using LipofectamineTM 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), in accordance with the manufacturer’s instructions. The efficiency of the knockdown was determined using reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Real-time RT-PCR analysis
The total RNA was extracted from treated and cultured PCa cells. Then, cDNA was synthesized from 500 ng RNA of each sample, 2 µL PrimeScript RT Master Mix (Takara Bio, Otsu, Japan), and RNase-free dH2O (Takara Bio, Otsu, Japan), which was added to a final volume of 10 µL. The total volume (10 µL) of each PCR reaction mixture contained 2.5 µL SYBR Green Supermix (Takara Bio, Otsu, Japan), 1 µL RNase-free dH2O, 1 µL cDNA, and 10 µM of each of forward and reverse primers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. The sequences of primers are summarized in Table 1.
Table 1
| Gene | Forward | Reverse |
|---|---|---|
| KMT2D | 5'-ACCATGTGAAGAACAGGAAGAG-3' | 5'-TCACCCTGGCTCAGATTAGA-3' |
| PPARγ | 5'-GACAGGAAAGACAACAGACAAATC-3' | 5'-GGGGTGATGTGTTTGAACTTG-3' |
| FASN | 5'-CAGGCACACACGATGGAC-3' | 5'-CGGAGTGAATCTGGGTTGAT-3' |
| ACC | 5'-CGGACTTCGGCAGAGGTAG-3' | 5'-TGCTCTGAAATTGCCTTGG-3' |
| ACLY | 5'-GCTGGTCCACATGAACAGG-3' | 5'-GCCTTCTGGATATTCAGGACTTT-3' |
| GAPDH | 5'-CGGATTTGGTCGTATTGGG-3' | 5'-CTGGAAGATGGTGATGGGATT-3' |
| SREBP1 | 5'-CGGAACCATCTTGGCAACAGT-3' | 5'-CGCTTCTCAATGGCGTTGT-3' |
| SREBP2 | 5'-CCTGGGAGACATCGACGAGAT-3' | 5'-TGAATGACCGTTGCACTGAAG -3' |
Oil red O (ORO) staining
The lipid concentration of neutral triglycerides and cholesterol esters were analyzed by ORO staining (MA0120, Meilunbio, China). PC-3 and DU-145 cells were seeded at 2×105 cells/well in a six-well plate. Each group of cells was equipped with three complex pores. The cell density was increased to approximately 80–95% to meet the staining standard. The cell medium was discarded, and the cells were gently rinsed with 10% phosphate-buffered saline (P1020, Solarbio, China). Paraformaldehyde (Beyotime) was used to fix the cells in each well at room temperature. The samples were then washed twice with deionized water. Approximately, 2 mL of ORO working solution was added to each well, and the cells were stained for approximately 1.5 h. A pipette gun was used to remove the ORO working fluid. After staining, the cells were washed thrice with deionized water. Intracellular staining images were captured using a microscope (Leica, Germany). After photographing, the deionized water was removed from the six-well plate. Finally, 500 µL of 100% isopropyl alcohol (Beyotime) was added to each well of the 96-well plate, and the optical density of each well was measured at 520 nm.
BODIPY 493/503 staining
We used the BODIPY 493/503(GC42959, GLPBIO, USA) to facilitate quantification of neutral lipid content. PC-3 and DU-145 cells were seeded at 2×105 cells/well in a six-well plate. Each group of cells was equipped with three complex pores. The cell density was increased to approximately 80–95% to meet the staining standard. Then, we stained and performed fluorescence photography according to the kit’s method.
The ProteinSimple analysis and Western blotting analysis
After treatment, the total protein was extracted on ice, as described previously (15). The ProteinSimple analysis (also called western immunoassay, Wes) (San Jose CA, USA) was performed for this study to analyze the expression levels of proteins, as described previously (18). All procedures were performed using the reagents in accordance with the manufacturer’s instructions. Briefly, 2 µL 5× fluorescent mixture was mixed with 8 µL protein lysate, then diluted to a concentration of 0.5–1 µg/µL, and finally heated at 95 ℃ for 5 min. The chemiluminescence substrates were distributed into the holes of the specified enzyme plate to avoid the formation of bubbles during the process. The enzyme plate was loaded into the instrument, and proteins were inhaled into a 14 or 25 capillary cartridge supplied by the manufacturer (Jess/Wes Separation 12–230 kDa 8×14 or 8×25 capillary cartridge kit). Using default settings, protein separation and immunodetection were automatically performed in individual capillaries. The obtained data were analyzed using the COMPASS software (ProteinSimple, San Jose, CA, USA). The peak intensity of the target protein (area under the curve) was normalized to that of the vinculin peak. Primary antibodies, which included FASN (#3180, 1:1,000), acetyl-CoA carboxylase (ACC, #3676, 1:1,000), and ATP citrate lyase (ACLY, #13390, 1:1,000), were purchased from Cell Signaling Technologies, and GAPDH was purchased from Santa Cruz Biotechnology.
Co-immunoprecipitation (Co-IP) assay
After PC-3 or DU-145 cells were extracted, samples were then incubated with the anti-KMT2D (Sigma, #HPA035977) or a control normal rabbit immunoglobulin G antibody overnight at 4 ℃ on a rolling platform. Protein A beads (Bimake, USA) were added to the sample for 2 h at 4 ℃; then the sample was washed six times using a washing buffer (Bimake, USA). The target protein was eluted from these beads using a 2× sodium dodecyl-sulfate (SDS) polyacrylamide gel electrophoresis loading buffer for 5 min. The IP samples were detected by western blotting. KMT2D was validated by electrophoresis in 6% SDS polyacrylamide (Meilunbio, China); 10% SDS polyacrylamide (Meilunbio, China) was used for other proteins. The proteins were then transferred on to a polyvinylidene difluoride membrane (0.2 µm) (Millipore, Bedford, USA). The primary antibodies used included KMT2D (Sigma, #ABE1867), KMT2D (Santa Cruz, #sc-293217), PPARγ (CST, #2443S), and GAPDH (Santa Cruz, #sc-47724).
DNA pull-down assay
Biotinylated PPAR response element (PPRE) DNA fragments were prepared using PCR and biotinylated primers by GZYXBIO Company (Guangzhou, China). DNA pull-down assay was performed as follows: magnetic beads bound to labeled DNA streptavidin were incubated with nuclear proteins overnight, and the bead-probe-protein complex was washed with 1 mL wash buffer. Next, the magnetic beads were resuspended in 200 µL 1× SDS loading buffer and denatured at 95 ℃ for 10 min. Finally, the primary antibodies were added, which included KMT2D (Proteintech, #27266-1-AP) and PPARγ (CST, #2443S). The PPRE region motif search was conducted using the MEME website (https://meme-suite.org/meme/). MEME is one of the most widely used tools for discovering new transcription factor-binding sites and protein domains.
Wound healing assay
On the first day, PC-3 cells were seeded in a six-well culture dish. On the following day, the cells were transfected with siRNA using LipofectamineTM 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), in accordance with the manufacturer’s instructions. Twenty-four hours after transfection, the cells were cultured to 100% fusion, and then tested against the control group. The extent of wound closure was calculated using ImageJ. Data are shown as the mean ± standard deviation (SD) of three independent experiments.
Cell viability assay
The PC-3 control and PC-3 KMT2D-knockdown cells were placed in a 96-well plate and treated with 10 µM ROSI (Sigma; St. Louis, MO, USA) or DMSO (Sigma; St. Louis, MO, USA). The cell viability assay was performed using a cell counting kit (CCK-8) (Fdbio Science, China), in accordance with the manufacturer’s instructions. The absorbance of the sample was measured at 450 nm.
Bioinformatics analysis
Gene expression of different sample types (496 PCa tissues and 52 normal prostate tissues) was obtained using the transcriptome data provided by The Cancer Genome Atlas (TCGA) database. Data of GSE60396 and GSE38341 were obtained from the GEO database. The Limma package (version: 3.40.2) of R software was used to study the differential gene expression of mRNAs. Spearman correlation analysis was used to analyze KMT2D expression and the typical gene expression of the lipid metabolism pathway. The correlation map was displayed using the R software package heatmap. Enrichment analysis was performed using the R package clusterProfiler (version 3.14.3) to obtain the results of gene set enrichment. Mutations in KMT2D and PPARγ in PCa were analyzed using the Cbioportal website (https://www.cbioportal.org/). target genes of PPARγ were validated using CistromeMap (https://cistrome.org/db/). The immunohistochemical profiles of normal prostate tissue and PRAD (Prostate adenocarcinoma) were derived from the Human Protein Atlas (https://www.proteinatlas.org/).
Statistical analysis
Data from multiple experiments are expressed as the mean ± SD. All data were analyzed using statistical software, and statistical differences between two groups (P values) were calculated using a Student’s t-test. P values for more than three groups were calculated using GraphPad Prism 8.0 and one-way analysis of variance. P<0.05 indicated significant difference.
Results
KMT2D knockdown in PCa cells inhibits lipogenesis
In our previous study (15), the KMT2D expression in 46 PCa tissues was higher than that in normal tissues, and high levels of KMT2D are associated with poor disease prognosis. In this study, we first validated the KMT2D expression in the GSE60396 and GSE38341 PCa datasets, and the results supported our previous study (Figure 1A). The protein expression levels of KMT2D and PPARG were collected from the immunohistochemical staining results of the Human Protein Atlas database (Table S1), and several representative pictures of KMT2D and PPARG in normal prostate tissues and PCa tissues were selected and listed in Figure 1B. Overall, in these tissues, the levels of the expression of KMT2D and PPARG proteins were higher in tumor tissues than in normal tissues. Then, we knocked down KMT2D significantly in the human PCa cell lines PC-3 and DU-145 (Figure 1C). ORO is a lysochrome and a fat-soluble dye used to stain triglycerides and lipoproteins (19). It is difficult to ionize, which renders it highly soluble in lipids. The PCa cells were then grouped for ORO staining and relative quantification (Figure 1D). Moreover, BODIPY 493/503 has specific recognition of neutral lipid content, and we further stained and verified the changes in lipid droplet in these two PCa cell lines (Figure 1E). The results demonstrated that KMT2D knockdown significantly reduced the lipid droplet content of PCa cells. Therefore, we hypothesized that KMT2D and lipid metabolism are linked to PCa.
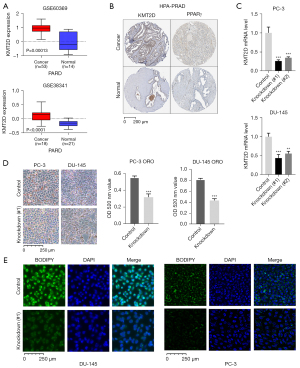
KMT2D knockdown inhibits the expression of genes related to lipid metabolism in PCa
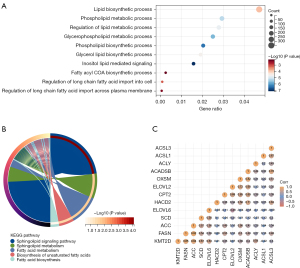
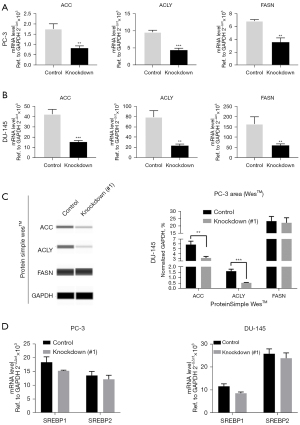
Here, PCa samples from TCGA database were used for bioinformatics analysis, we compared the differential genes in patients with high- and low-expression of KMT2D (cut of 50%) and performed KEGG and GO functional enrichment analysis, which identified many significant pathways, including lipid-related pathways and fatty acid metabolism-related pathways (Figure 2A,2B). Among them, 11 genes were enriched in the fatty acid metabolic pathway (P<0.05). Correlation analysis showed that KMT2D expression was significantly correlated with FASN, ACC, SCD, and ACLY expression (Figure 2C). To further demonstrate the effect of KMT2D on lipogenesis, the mRNA levels of ACC, ACLY, and FASN after KMT2D knockdown were quantified in PC-3 and DU-145. The results showed a significant decrease in transcript levels (Figure 3A,3B). In addition, we also analyzed the changes in lipid metabolism-related protein levels after KMT2D knockdown in PC-3 cells. These results showed that both ACC and ACLY protein levels were downregulated after KMT2D knockdown for 48 h (Figure 3C). Apparently, KMT2D affected lipid metabolism-related genes in PCa. According to related reports, sterol regulatory-element binding protein 1/2 (SREBP1/2) and PPARγ are important protein regulatory centers in lipid metabolism (20,21). However, we further detected that there was no significant change in SREBP1/2 with the knockdown of KMT2D (Figure 3D).
Critical roles of KMT2D in PPARγ transcriptome-induced changes in PCa cells
Although our study did not find that KMT2D affected the transcript levels of SREBP1/2, and some studies did not detect the interaction of the KMT2D complex with SREBP1 and SREBP2 (16,22). Therefore, we speculated that KMT2D affected lipogenesis in PCa, possibly through the PPARγ pathway. Here, ROSI, a PPARγ synthesis agonist, was used to initially explore the role of KMT2D upstream or downstream of the PPARγ lipid metabolism pathway. After ROSI treatment for 24 h (10 µM), the control group (PN) showed a significant increase in the lipid droplet content compared with the knockdown group (PK) (P<0.001). Additionally, the lipid droplet content in the knockdown group after ROSI treatment (PK-ROSI) was not significantly different from that in the control group after DMSO treatment (PN-DMSO) (P>0.05, Figure 4A). Further analysis of the transcriptional expression level of PPARγ, we found that compared with PN-DMSO cells, ROSI or DMSO treatment made the activities of PN-ROSI, PK-DMSO and PK-ROSI become 2.5, 0.5, 0.7 times (PN-DMSO expression level is set to 1) (Figure 4B). The ROSI cannot effectively induce the increase of PPARγ expression after KMT2D knockdown. These results indicate that KMT2D knockdown reduced the agonistic effect of ROSI on lipid synthesis in the PPARγ pathway. Thus, KMT2D plays an important role in PPARγ transcriptional activation. Additionally, after ROSI treatment of PC-3 cells in the knockdown group, the mRNA levels of ACC, ACLY, and FASN and the protein levels of PPARγ, ACC, and ACLY were not as high as those in the control group after ROSI treatment (P<0.05, Figure 4C,4D).
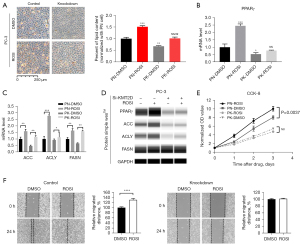
As KMT2D knockdown caused disturbances in the lipid metabolism of cells, we speculated that this is related to the cancer-promoting effect of KMT2D in PCa. The CCK-8 cell proliferation analysis revealed that the proliferation activity in ROSI-treated cells was significantly higher than that in the DMSO-treated cells in the control group (P<0.05, Figure 4E). However, in the KMT2D knockdown group, no significant difference was found in the proliferation activity between the ROSI-treated and DMSO-treated cells (P>0.05). Next, the effect of ROSI on the migration ability of PC-3 control and KMT2D knockdown cells was evaluated (Figure 4F); the results revealed that ROSI promoted cell migration in the control group (P<0.05), whereas KMT2D knockdown cells did not show a significant difference in cell migration.
The PPARγ-KMT2D complex is a key regulator for PCa lipid metabolism
In the study of hepatic steatosis, BL1 kinase binds KMT2D to PPARγ to form the KMT2D-PPARγ complex, where KMT2D plays a demethylation regulatory role (16). Adipogenesis and hepatic steatosis share a similar gene regulatory program (17). Combined with above findings, our finding suggests that there may be similar KMT2D-PPARγ binding in PCa. In addition, Mutually-exclusive tumor-driver genes often have the same Pathway. We analyzed the genetic variation in the cohort of PCa patients by using cBioPortal, and found that even though the total mutation ratio of KMT2D and PPARγ was only 6% and 1%, interestingly, the mutations of PPARγ and KMT2D in the cohort of PCa presented obvious “mutual exclusion” (Figure 5A).
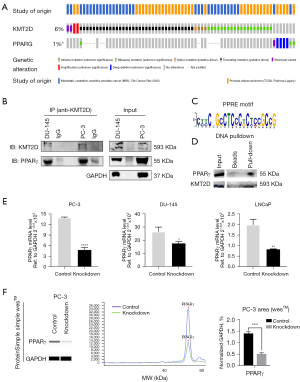
To verify this hypothesis, the Co-IP assay was performed using PC-3 and DU-145 cells to further determine the underlying mechanism (Figure 5B). We then performed DNA pull-down experiments and still found PPARγ and KMT2D present in the pull-down protein by western blotting (Figure 5C,5D). The findings suggest that PPARγ acted as a transcription factor that recruited KMT2D to its target genes. In the above-mentioned study on the KMT2D-PPARγ complex (16), it was also proposed that KMT2D induces PPARγ expression because PPARγ itself is a target of the PPARγ-KMT2D axis. Therefore, to further verify whether there was a similar mechanism in PCa, the PPARγ mRNA and protein levels of PC-3, DU-145, and LNCaP cells after KMT2D knockdown were examined (Figure 5E,5F). The results showed that PPARγ expression was downregulated when KMT2D was knocked down.
Discussion
Because androgen receptor (AR) is the main driver of PCa, some patients with advanced PCa who cannot be cured by surgery or radiation mainly opt for endocrine therapy targeting androgens or ARs. The progression to castration-resistant PCa is often inevitable, which requires a combination of anti-androgenic drugs such as enzalutamide and abiraterone. Furthermore, from a metabolic point of view, men treated with ADT experience increased obesity and metabolic syndrome, with an increase in the total cholesterol, triglyceride, and high-density lipoprotein concentrations (23). Thus, disease progression is significantly associated with lipid-specific changes in patients with PCa. In the treatment of advanced PCa, there is an urgent need to identify cellular pathways that allow tumors to escape anti-androgen therapy. Studies have shown that a high-fat diet plays an important role in PCa progression (21,24,25). Lipid metabolism rebinds the PCa metabolome to support growth and resistance to various endocrine therapy drugs; however, the exact mechanism remains unclear. Therefore, further studies on lipid metabolism in PCa may provide new therapeutic opportunities for patients with advanced PCa. Combined with our above study, we speculated and confirmed that KMT2D participates in the PCa lipid metabolism pathway and plays an important role in regulating transcription.
In the presence of ligands, the PPARγ-retinoid X receptor (RXR) complex will be activated in the nucleus, bind to the PPRE sequence of the target gene promoter, and participate in the regulation of the transcription of the downstream target gene. However, the mechanism of this process after PPARγ enters the nucleus needs to be further explored. Related research has shown that when PPARγ is activated, it suppresses the growth of pancreatic, biliary, oral, esophageal, gastric, and colorectal tumor cells (26). Paradoxically, PPARγ plays a role in promoting PCa progression. For example, increased PPARγ and cutaneous fatty acid-binding protein expression levels are associated with an increase in the malignant phenotype of PCa cells (27). PPARγ may also be used as a modulator of AR, and inhibiting PPARγ expression leads to a decline in PCa growth and proliferation (28). Our research further supports the hypothesis that PPARγ is an oncogene, which is also the mainstream view. After KMT2D knockdown, a decrease in the PPARγ level, expression of lipid metabolism pathway downstream genes, and the lipid droplet content in PCa cells was observed. Even with low-dose ROSI treatment, PC-3 cells with KMT2D knockdown could not fully recover their proliferation and migration viability.
As a ligand of PPARγ, ROSI binds to PPARγ and causes conformational changes in PPARγ. Regardless of the concentration, ROSI stimulates apoptotic factors, such as p53 and PTEN, and inhibits anti-apoptotic factors, such as BCL-2/BCL-XL, in a PPARγ-independent manner to induce cell apoptosis (26). However, low ROSI concentrations (5–10 µM) induce the activation of PPARγ, increase lipid droplet production and energy supply (29), and reduce or even offset cancer cell apoptosis. In this study, we found that ROSI activated PPARγ to increase the lipid droplet content in PC-3 cells. However, after KMT2D knockdown, the effect of ROSI treatment became limited, which illustrated the effect of KMT2D on the activation of the PPARγ pathway (Figure 4A-4C). It is worth noting that the protein level of FASN, which did not fluctuate significantly in the four groups, and its mRNA level after ROSI treatment was nearly the same in the knockdown and control groups. We hypothesize that there are two reasons for this. First, several studies have reported that the correlation between PPARγ and FASN is abnormally significant (30,31). Although the PPARγ levels decrease after KMT2D knockdown, the remaining levels promote the normal translation of FASN. Second, as an important lipid synthesis gene, FASN might be regulated by core lipid metabolism proteins, such as PPARγ and SREBP. Therefore, after the downregulation of the PPARγ pathway, other pathways receive feedback and supplement FASN protein levels.
Because AR itself regulates fatty acid metabolism, there may be cross-regulation of some adipogenic genes in AR-positive PCa cells. However, PC-3 and DU-145 cells are AR-negative, which allows the use of these cells to further clarify that, in addition to AR genes, PPARγ also directly controls adipogenic genes to regulate PCa cell growth (32). However, as a 593 kDa super large protein complex, KMT2D presents many challenges in the selection of antibodies and the exploration of its mechanism. Through Co-IP analysis, PPARγ and KMT2D bound to each other to regulate the transcription of PPARγ downstream target genes in PCa cells. Chromatin immunoprecipitation sequencing (CHIP-seq) was used to analyze whether there was a KMT2D peak in the region of the PPARγ-binding site. However, even though this technology was familiar, we had to rely on bioinformatics analysis because we could not find suitable antibodies and conditions conducive to effective CHIP. Based on the analysis of the Meme website, the motif of PPRE was determined, and DNA pulldown analysis was performed, which demonstrated once again that PPARγ and KMT2D bound to each other in the promoter regions of PPARγ target genes and had a role in transcriptional regulation. Overall, these two pieces of data indicated that PPARγ, as a major transcription factor in lipid metabolism, recruited KMT2D to target genes (Figure 5B-5D). As H3K4 methylation is presumably associated with gene activation (33), it will be particularly interesting to investigate the roles of KMT2D and the potential synergy of the associated H3K4 methyltransferase activities and regulation of the PPARγ pathway during adipogenesis. However, this hypothesis has not been confirmed, and detailed discussion with other researchers is necessary in the future.
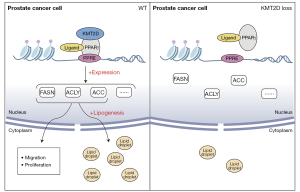
In summary, the key link between PCa lipid metabolism and its therapeutic goals is to identify upstream regulators that affect tumor metabolism (34). Our study showed that KMT2D participated in the upstream pathway of PPARγ lipid metabolism to regulate the transcription and translation of PPARγ and downstream genes associated with lipid metabolism and affected lipid synthesis in cancer cells (Figure 6). ROSI induces apoptosis in androgen-dependent and hormone-refractory tumors (26). However, clinical studies have shown that ROSI is ineffective as monotherapy for PCa treatment (35). Researchers have proposed that imatinib may antagonize KMT2D and PPARγ binding (16), but it has not been discussed in depth in PCa, and drug development targeting lipid metabolism in advanced PCa will require time.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Funding: This work was supported by the Natural Science Foundation of Guangdong Province of China (No. 2021A1515011023) and the National Natural Science Foundation of China (No. 81502577).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-431/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-431/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-431/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-431/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Rebello RJ, Oing C, Knudsen KE, et al. Prostate cancer. Nat Rev Dis Primers 2021;7:9. [Crossref] [PubMed]
- Buszewska-Forajta M, Pomastowski P, Monedeiro F, et al. Lipidomics as a Diagnostic Tool for Prostate Cancer. Cancers (Basel) 2021;13:2000. [Crossref] [PubMed]
- Kuemmerle NB, Rysman E, Lombardo PS, et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol Cancer Ther 2011;10:427-36. [Crossref] [PubMed]
- Dang Q, Chen YA, Hsieh JT. The dysfunctional lipids in prostate cancer. Am J Clin Exp Urol 2019;7:273-80. [PubMed]
- Butler LM, Mah CY, Machiels J, et al. Lipidomic Profiling of Clinical Prostate Cancer Reveals Targetable Alterations in Membrane Lipid Composition. Cancer Res 2021;81:4981-93. [Crossref] [PubMed]
- Wang X, Kruithof-de Julio M, Economides KD, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009;461:495-500. [Crossref] [PubMed]
- Lai B, Lee JE, Jang Y, et al. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 2017;45:6388-403. [Crossref] [PubMed]
- Fagan RJ, Dingwall AK. COMPASS Ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer Lett 2019;458:56-65. [Crossref] [PubMed]
- Li J, Xu C, Lee HJ, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020;580:93-9. [Crossref] [PubMed]
- Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer 2015;15:334-46. [Crossref] [PubMed]
- Schwenty-Lara J, Nehl D, Borchers A. The histone methyltransferase KMT2D, mutated in Kabuki syndrome patients, is required for neural crest cell formation and migration. Hum Mol Genet 2020;29:305-19. [Crossref] [PubMed]
- Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015;21:1190-8. [Crossref] [PubMed]
- Mendiratta G, Ke E, Aziz M, et al. Cancer gene mutation frequencies for the U.S. population. Nat Commun 2021;12:5961. [PubMed]
- Lv S, Ji L, Chen B, et al. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene 2018;37:1354-68. [Crossref] [PubMed]
- Kim DH, Kim J, Kwon JS, et al. Critical Roles of the Histone Methyltransferase MLL4/KMT2D in Murine Hepatic Steatosis Directed by ABL1 and PPARγ2. Cell Rep 2016;17:1671-82. [Crossref] [PubMed]
- Okumura T. Role of lipid droplet proteins in liver steatosis. J Physiol Biochem 2011;67:629-36. [Crossref] [PubMed]
- Du H, Wang Y, Zeng Y, et al. Tanshinone IIA Suppresses Proliferation and Inflammatory Cytokine Production of Synovial Fibroblasts from Rheumatoid Arthritis Patients Induced by TNF-α and Attenuates the Inflammatory Response in AIA Mice. Front Pharmacol 2020;11:568. [Crossref] [PubMed]
- Wang Y, Goulart RA, Pantanowitz L. Oil red O staining in cytopathology. Diagn Cytopathol 2011;39:272-3. [Crossref] [PubMed]
- Galbraith L, Leung HY, Ahmad I. Lipid pathway deregulation in advanced prostate cancer. Pharmacol Res 2018;131:177-84. [Crossref] [PubMed]
- Chen M, Zhang J, Sampieri K, et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet 2018;50:206-18. [Crossref] [PubMed]
- Lee JE, Wang C, Xu S, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2013;2:e01503. [Crossref] [PubMed]
- Saylor PJ, Karoly ED, Smith MR. Prospective study of changes in the metabolomic profiles of men during their first three months of androgen deprivation therapy for prostate cancer. Clin Cancer Res 2012;18:3677-85. [Crossref] [PubMed]
- Huang M, Narita S, Koizumi A, et al. Macrophage inhibitory cytokine-1 induced by a high-fat diet promotes prostate cancer progression by stimulating tumor-promoting cytokine production from tumor stromal cells. Cancer Commun (Lond) 2021;41:389-403. [Crossref] [PubMed]
- Kwan HY, Liu B, Huang C, et al. Signal transducer and activator of transcription-3 drives the high-fat diet-associated prostate cancer growth. Cell Death Dis 2019;10:637. [Crossref] [PubMed]
- Yousefnia S, Momenzadeh S, Seyed Forootan F, et al. The influence of peroxisome proliferator-activated receptor γ (PPARγ) ligands on cancer cell tumorigenicity. Gene 2018;649:14-22. [Crossref] [PubMed]
- Forootan FS, Forootan SS, Malki MI, et al. The expression of C-FABP and PPARγ and their prognostic significance in prostate cancer. Int J Oncol 2014;44:265-75. [Crossref] [PubMed]
- Tew BY, Hong TB, Otto-Duessel M, et al. Vitamin K epoxide reductase regulation of androgen receptor activity. Oncotarget 2017;8:13818-31. [Crossref] [PubMed]
- Moss PE, Lyles BE, Stewart LV. The PPARγ ligand ciglitazone regulates androgen receptor activation differently in androgen-dependent versus androgen-independent human prostate cancer cells. Exp Cell Res 2010;316:3478-88. [Crossref] [PubMed]
- Ahmad I, Mui E, Galbraith L, et al. Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer. Proc Natl Acad Sci U S A 2016;113:8290-5. [Crossref] [PubMed]
- Guo J, Zhang S, Fang L, et al. In utero exposure to phenanthrene induces hepatic steatosis in F1 adult female mice. Chemosphere 2020;258:127360. [Crossref] [PubMed]
- Elix CC, Salgia MM, Otto-Duessel M, et al. Peroxisome proliferator-activated receptor gamma controls prostate cancer cell growth through AR-dependent and independent mechanisms. Prostate 2020;80:162-72. [Crossref] [PubMed]
- Mansour M, Schwartz D, Judd R, et al. Thiazolidinediones/PPARγ agonists and fatty acid synthase inhibitors as an experimental combination therapy for prostate cancer. Int J Oncol 2011;38:537-46. [Crossref] [PubMed]
- Stoykova GE, Schlaepfer IR. Lipid Metabolism and Endocrine Resistance in Prostate Cancer, and New Opportunities for Therapy. Int J Mol Sci 2019;20:2626. [Crossref] [PubMed]
- Hatton JL, Yee LD. Clinical Use of PPARgamma Ligands in Cancer. PPAR Res 2008;2008:159415. [Crossref] [PubMed]


