
Clinicopathological significance and prognostic value of polypyrimidine tract binding protein 1 (PTBP1) in gastric cancer
Introduction
Gastric cancer (GC) is one of the leading causes of cancer-related death in the world, especially in Asia (1,2). In recent years, advances in diagnostic and therapeutic technologies such as surgical techniques and novel targeted molecular agents, remarkably have improved prognosis of gastric cancer patients. However, invasion and metastasis are still the primary causes of poor outcome in patients with gastric cancer (2,3). Thus, it is essential to search for new molecular biomarkers related to gastric cancer prognosis and metastasis.
Polypyrimidine tract binding protein 1 (PTBP1) (also known as hnRNPI and PTB), was expressed by nearly all types of cells, including CD4+ T cells, myoblasts, neuronal progenitor cells and cardiomyocytes (4-8). As a member of heterogeneous nuclear ribonucleoprotein (hnRNP) family, PTBP1 was assumed to be a family of RNA-binding proteins (RBPs) (9), which mediated interactions of disparate RNAs and controlled post-transcriptional regulation of gene expression, playing an essential function in regulating RNA metabolism (10,11). An increasing number of researches indicated PTBP1 was involved in many processes of post-transcriptional regulation of gene expression, such as modification, alternative splicing, export, localization, stability, polyadenylation, translation and degradation of mRNA (9,10,12). Recently, PTBP1 has been reported to play important roles in the progression of malignancies. For example, in breast cancer, PTBP1 was highly expressed and contributed to cell invasion and metastasis (13). In colon cancer, high expression of PTBP1 promoted invasion by alternative splicing of cortactin (14), moreover, PTBP1 knockdown enhanced the sensitivity of drug-resistant colon cancer cells to vincristine and oxaliplatin (15). Additionally, PTBP1 was also reported to be involved in tumor growth and invasion in diverse tumors, including bladder cancer (16), clear-cell renal carcinoma (17), hepatocellular carcinoma (18), lung carcinoma (19) and glioma (20). As for gastric cancer, Sugiyama et al. (21) found that PTBP1 expressed higher and mediated the function of miR-133b on cancer cell invasion and metastasis by acting as a target gene of miR-133b. However, the clinical significance and prognostic value of PTBP1 in gastric cancer have not been sufficiently elucidated.
In the present study, bioinformatic analysis, reverse transcription polymerase chain reaction (RT-PCR) and immunohistochemical staining were performed to detect the expression type of PTBP1 in gastric cancer at mRNA and protein level. Association of PTBP1 protein expression with clinicopathological features and overall survival was analyzed furthermore. In addition, the biological role of PTBP1 in gastric cancer proliferation, migration and invasion was explored in vitro. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-303/rc).
Methods
Bioinformatics analysis
The expression levels of PTBP1 mRNA in gastric cancer patients and association of PTBP1 expression with overall survival were obtained from The Cancer Genome Atlas (TCGA) database (https://TCGA-data.nci.nih.gov/) (data set name: phs000178). R software (ver. 3.6.3) was used to analyze the data above. Visualization was performed by R language (ggplot2 mainly).
Patients and sample collection
A total of 311 cases of gastric cancer tissues and corresponding 30 adjacent non-tumor tissues, were obtained from patients (220 males and 91 females), who had undergone initial surgical treatment, with a median age of 58 years (range, 25–88 years) between January 2002 and October 2006 at the First Affiliated Hospital of Sun Yat-sen University. Located at 5 cm from the tumor margin and diagnosed as non-atrophic gastritis, the adjacent non-tumor tissues, were morphologically normal. Patients were not previously received chemotherapy, radiotherapy or other surgeries, followed up from 1 to 62 months. The definitions of gastric cancer histopathology and clinical stage were according to the 8th edition of the TNM Staging system of American Joint Committee on Cancer (AJCC). Unabridged clinicopathological data were acquired from medical records. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Review Committee of the Affiliated Hospital of Sun Yat-sen University (No. 2022-185) and individual consent for this retrospective analysis was waived.
Tissue microarray and immunohistochemical staining and scoring
Tissue cores from gastric cancer tissues (1.0–1.5 mm in diameter) were punched and removed. Tissue microarray (TMA) was constructed by transferring the removed tissue cores to a recipient paraffin mass devised beforehand and pre-punched. Cut from the TMA, paraffin tissue chips were dewaxed and hydrated and then boiled for 3–4 min in a pressure cooker. Endogenous peroxidases were inactivated by 3% hydrogen peroxide solution for 40 min. Subsequently, non-specific antigens were blocked with 5% goat serum for 30 min. TMA sections were incubated with a primary antibody against PTBP1 (1:150; Abcam) at 4 ℃ overnight. Secondary antibody used was anti-rabbit IgG (Dako). The TMA slices were counterstained with hematoxylin, sequentially dehydrated and vitrified. Eventually the TMA segments were sealed with neutral resins for long-term preservation. Intensity of immunostaining was scored as 1 (no immunostaining), 2 (weak immunostaining), 3 (moderate immunostaining), and 4 (strong immunostaining). The proportion of immunoreactive tumor cells was scored as 1 (0–24%), 2 (25–49%), 3 (50–74%) and 4 (75–100%). Staining index was calculated as multiples of staining intensity and staining percentage, ranging from 1 to 16). Low expression was defined as a staining index <8 and high expression with a staining index ≥8. Two professional pathologists were blinded to the clinical outcome and employed to assess the specimens independently.
Cell culture
All cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HGC-27, MKN-28, MKN-45 and SGC-7901 cell lines were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin. AGS and GES-1 cell lines were cultivated in Dulbecco’s modified eagle medium (DMEM) (Gibco) containing 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37 ℃ in a humidified atmosphere with 5% CO2.
Quantitative real-time PCR
Total RNA was isolated using Trizol reagent (ACCURATE BIOLOGY AG, China) from cells and reverse-transcribed to cDNA with an Evo M-ML V RT Kit (ACCURATE BIOLOGY AG, China). The reaction program was performed as below: 1 cycle of 37 ℃ for 15 min, 85 ℃ for 5 sec and 4 ℃ for 10 min. The sequences of primers were as follows (5'–3'): PTBP1 (AGGCGGTGAACTCGGTCCAG and TGCAGCAGGCGTTGTAGATGTTC); GAPDH (GACTCATGACCACAGTCCATGC and GAGGCAGGGATGATGTTCTG). Quantitative real-time PCR (qRT-PCR) was performed using the SYBR® Green Premix Pro Taq HS qPCR Kit (ACCURATE BIOLOGY AG, China), conducted as follows: 95 ℃ for 30 sec, followed by 40 cycles of 95 ℃ for 5 sec and 60 ℃ for 30 sec. The relative expression level of genes was calculated using the 2−ΔΔCq method.
Western blot
Proteins were extracted from cells by radio immunoprecipitation assay (RIPA) buffer and quantified using bicinchoninic acid assay. Equal protein amounts were loaded on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and followed by transferred to a polyvinylidene fluoride (PVDF) membrane. After blocking with 5% skimmed milk, membranes were incubated with anti-PTBP1 (1:10,000; Abcam) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:10,000; Abcam) at 4 ℃ overnight, followed by goat anti-rabbit IgG (1:5,000; Abcam) at room temperature for 1 h. Protein bands were visualized by the enhanced chemiluminescence kit.
Cell counting kit-8 (CCK-8) assay
Proliferation activity of cells was detected by CCK-8 assay. In total, 100 µL of cell suspension containing 2×103 cells were added into 96-well plates. Cells were cultured for 0, 24, 48, 72 and 96 h respectively at 37 ℃ and 5% CO2. CCK-8 solution (10 µL) was put into each well and incubated for 3 h. The absorbance was measured at a wavelength of 450 nm. The above experiment was repeated for three times.
Transwell migration and invasion assay
Migration and invasion assay were performed using Transwell chambers. After transfection, 1×104–10×104 cells were seeded into the upper chambers of the inserts cultured with 200 µL FBS-free medium. Then, 600–800 µL medium containing 20% FBS was added to the lower chamber. For invasion assay, top chamber had been coated with 50–60 µL diluted Matrigel at 37 ℃ for 6 h before cells were seeded. After incubation for 48 h, cells were fixed by 3% paraformaldehyde and stained with 0.1% crystal violet solution. Cells on the upper surface of the membrane were wiped away. Five randomly chosen fields were quantitatively analyzed by counting the mean number of cells via a light microscope at ×100 magnification. The assays were performed in triplicates.
Statistical analyses
Data were analyzed for descriptive statistics using SPSS statistical software version 24.0 (SPSS Inc. Chicago, IL, USA). Kaplan-Meier analysis with log-rank test was used for survival analysis. The Cox proportional-hazards regression method was employed for multivariate analysis. We used Pearson’s chi-squared test or Fisher’s exact test for comparing differences in categorical variables. P<0.05 was considered statistically significant.
Results
Bioinformatics analysis of PTBP1 expression and prognostic value in gastric cancer
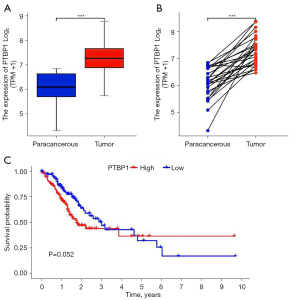
Data of mRNA expression and clinical information from 375 cases of gastric cancer and 32 cases of corresponding adjacent tissues were acquired from TCGA database (data set name: phs000178). The differential expression analysis of PTBP1 between gastric cancer and paracancerous tissues revealed that PTBP1 expression was significantly higher in gastric cancer tissues than that in adjacent tissues (P<0.001) (Figure 1A,1B). Using R language, the association between PTBP1 expression and prognosis was analyzed, showing that there was no statistic difference between PTBP1 expression and prognosis in gastric cancer patients (P=0.052), but there was a trend that high level of PTBP1 predicted poor prognosis (Figure 1C).
Expression of PTBP1 in gastric cancer cells and tissues
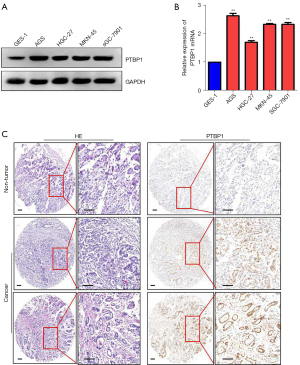
To confirm the results from TCGA data, RT-PCR and western blot were used to examine the expression of PTBP1 in gastric cancer cell lines (AGS, HGC-27, MKN-45 and SGC-7901) and gastric mucosal epithelial cell line GES-1. We observed that PTBP1 expression was increased at mRNA and protein level in gastric cancer cell lines compared to GES-1 (P<0.05) (Figure 2A,2B), which was consistent with the results from TCGA database. We then chose 311 gastric cancer and 30 adjacent non-tumor tissue microarray and verified the expression of PTBP1 protein in gastric cancer tissues by immunohistochemistry (IHC). Results indicated that PTBP1 was mainly expressed in the nucleus of gastric cancer tissues. High expression of PTBP1 (staining index ≥8) was detected in 64.6% (201/311) of gastric cancer tissues, while low expression of PTBP1 was observed in all 30 adjacent non-tumor tissues. Typical IHC staining images for PTBP1 in gastric cancer tissues were shown in Figure 2C. These results prompted that PTBP1 expression was increased in gastric cancer.
Correlation between PTBP1 expression and clinicopathologic characteristics
To detect the clinical significance of PTBP1 in GC, the relationships between the expression of PTBP1 and clinicopathological parameters were analyzed. High or low expression rates of PTBP1 protein in GC with respect to several standard clinicopathological features are presented in Table 1. It was showed that PTBP1 protein overexpression was significantly related with gender, age at surgery, histological type, TNM stage and lymph node metastasis. The frequency of PTBP1 positivity was significantly higher in male patients compared with in female patients (68.6%, 151/220 vs. 54.9%, 50/91, P=0.022). The expression of PTBP1 was higher in patients ≥58 years old at surgery compared with patients <58 years old at surgery (70.8%, 119/168 vs. 57.3%, 82/143, P=0.013). Furthermore, the frequency of PTBP1 positivity was significantly higher in intestinal type gastric cancer compared with in diffuse type gastric cancer (74.0%, 74/100 vs. 60.2%, 127/211, P=0.017). The expression of PTBP1 was higher in gastric cancer with lymph node metastasis compared with those without lymph node metastasis (67.5%, 168/249 vs. 53.2%, 33/62, P=0.036). Moreover, the proportion of PTBP1 positive samples was lower in early TNM stage (I + II) than those in the advanced stage (III + IV) (53.4%, 31/58 vs. 67.2%, 170/253, P=0.048). There was no significant correlation between PTBP1 protein expression and the other clinicopathological parameters, such as tumor size and T classification (P>0.05).
Table 1
| Characteristics | Cases | PTBP1 expression | P valuea | |
|---|---|---|---|---|
| High | Low | |||
| Gender | 0.022* | |||
| Male | 220 | 151 (68.6%) | 69 (31.4%) | |
| Female | 91 | 50 (54.9%) | 41 (45.1%) | |
| Age at surgery | 0.013* | |||
| <58b | 143 | 82 (57.3%) | 61 (42.7%) | |
| ≥58 | 168 | 119 (70.8%) | 49 (29.2%) | |
| Tumor size | 0.738 | |||
| <5 cm | 160 | 102 (63.7%) | 58 (36.3%) | |
| ≥5 cm | 151 | 99 (65.5%) | 52 (34.5%) | |
| Histological type | 0.017* | |||
| Intestinal | 100 | 74 (74.0%) | 26 (26.0%) | |
| Diffuse | 211 | 127 (60.2%) | 84 (39.8%) | |
| T classification | 0.204 | |||
| T1–2 | 51 | 29 (56.9%) | 22 (43.1%) | |
| T3–4 | 260 | 172 (66.1%) | 88 (33.9%) | |
| TNM stage | 0.048* | |||
| I + II | 58 | 31 (53.4%) | 27 (46.5%) | |
| III + IV | 253 | 170 (67.2%) | 83 (32.8%) | |
| Lymph node metastasis | 0.036* | |||
| Present | 249 | 168 (67.5%) | 81 (32.5%) | |
| Absent | 62 | 33 (53.2%) | 29 (46.8%) | |
a, Chi square test; b, median age; *, P<0.05. PTBP1, polypyrimidine tract binding protein 1.
Association of PTBP1 expression with prognosis in gastric cancer patients
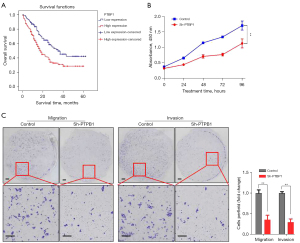
The relationship between PTBP1 expression and survival time in gastric cancer patients was analyzed by Kaplan-Meier analysis and log-rank test. A total of 261 cases with complete follow-up information were included. The results revealed that a significant impact of well-known clinicopathological prognostic features such as tumor size (P=0.041), histological type (P=0.016), T classification (P<0.001), lymph node metastases (P<0.001), and TNM stage (P<0.001) on the survival of gastric cancer patients. Furthermore, overall survival was significantly impaired in patients with high expression of PTBP1 compared to patients with low expression of PTBP1 in tumors (P=0.003) (Table 2) (Figure 3A). The median overall survival time was significantly shorter in high PTBP1 expression group (27.78±2.12 months) compared to low PTBP1 expression group (37.72±2.65 months) (Table 2).
Table 2
| Characteristics | Cases | Mean ± SE | Median ± SE | P value |
|---|---|---|---|---|
| Gender | 0.991 | |||
| Male | 181 | 31.64±2.09 | 25.00±3.18 | |
| Female | 80 | 32.11±3.02 | 21.00±4.38 | |
| Age at surgery | 0.538 | |||
| <58a | 121 | 32.82±2.48 | 26.00±2.46 | |
| ≥58 | 140 | 31.18±2.37 | 22.00±4.34 | |
| Tumor size | 0.041 | |||
| <5 cm | 131 | 34.87±2.39 | 31.00±6.81 | |
| ≥5 cm | 130 | 28.38±2.29 | 19.00±2.21 | |
| Histological type | 0.016 | |||
| Diffuse | 75 | 29.17±1.95 | 19.00±2.85 | |
| Intestinal | 186 | 39.38±3.31 | 25.00±2.51 | |
| T classification | <0.001 | |||
| T1–2 | 39 | 47.98±4.09 | NR | |
| T3–4 | 222 | 29.11±1.78 | 19.00±2.29 | |
| TNM stage | <0.001 | |||
| I + II | 48 | 46.11±4.15 | NR | |
| III + IV | 213 | 28.77±1.80 | 19.00±2.10 | |
| Lymph node metastasis | <0.001 | |||
| Present | 216 | 28.99±1.79 | 19.00±2.76 | |
| Absent | 45 | 48.27±3.89 | NR | |
| PTBP1 expression | 0.003 | |||
| Low | 94 | 37.72±2.65 | 35.00±6.28 | |
| High | 167 | 27.78±2.12 | 18.00±2.26 | |
a, median age. SE, standard error; NR, not reach.
Then, multivariate cox regression analysis showed histological type [hazard ratio (HR) =1.715, 95% confidence interval (CI): 1.103–2.667, P=0.017], lymph node metastasis (HR =3.857, 95% CI: 1.794–8.292, P=0.001), TNM stage (HR =3.743, 95% CI: 1.819–7.700, P<0.001) as well as PTBP1 expression (HR =1.763, 95% CI: 1.207–2.575, P=0.003) were identified as independent prognostic risk factors in patients with gastric cancer (Table 3).
Table 3
| Predictor | SE | Overall survival | ||
|---|---|---|---|---|
| HR | 95% CI | P value | ||
| Histological type (intestinal vs. diffuse) | 0.225 | 1.715 | 1.103–2.667 | 0.017 |
| Lymph node metastasis (present vs. absent) | 0.391 | 3.857 | 1.794–8.292 | 0.001 |
| TNM stage (I + II vs. III + IV) | 0.368 | 3.743 | 1.819–7.700 | <0.001 |
| PTBP1 expression (high vs. low) | 0.193 | 1.763 | 1.207–2.575 | 0.003 |
SE, standard error; HR, hazard radio; CI, confidence interval.
Effect of PTBP1 on gastric cancer cell proliferation, migration, and invasion
In order to explore the biological role of PTBP1 in gastric cancer, AGS cells, which were observed the highest PTBP1 protein expression, were selected for functional experiments. RT-PCR and western blot were performed to verify the knockdown efficiency of PTBP1 in AGS cells (Figure S1). CCK-8 assay was used to examine the effect of PTBP1 knockdown on cell proliferation. Through four days of continuous observation, the proliferation capacity of AGS cells with low expression of PTBP1 was significantly inhibited compared to the control group (P<0.001) (Figure 3B). Transwell assays were conducted to examine the migration and invasion ability. After PTBP1 expression had been suppressed, the migratory and invasive ability of AGS cells were remarkably reduced (P<0.001) (Figure 3C). Overall, these results demonstrated that PTBP1 might promote gastric cancer cell proliferation, migration and invasion, potentially leading to unfavorable prognosis of gastric cancer patients.
Discussion
In the present study, we confirmed that PTBP1 expression was increased in gastric cancer cells and tissues. We then revealed that PTBP1 protein expression was remarkably related to several clinicopathological features, including gender, age at surgery, histological type, TNM stage and lymph node metastasis. Moreover, high expression PTBP1 predicts poor prognosis in gastric cancer patients, and may be an independent prognostic factor for overall survival. Additionally, we determined that knockdown of PTBP1 substantially suppressed the proliferation, migration and invasion of gastric cancer cells in vitro.
Consistent with the results that PTBP1 was highly expressed in tumor tissues and predicted poor prognosis in diverse tumors, the present study demonstrated that PTBP1 expression was elevated at mRNA and protein level and may be served as an independent predictor of unfavourable prognosis in gastric cancer. Moreover, the association between PTBP1 and clinicopathological features in gastric cancer has been explored in detail. Although PTBP1 expression level has been detected in many types of tumors, data for the clinical significance of PTBP1 in cancers was limited. In breast cancer, high PTBP1 expression was correlated with positive human epidermal growth factor receptor 2 (her-2) expression, present lymph node metastasis, and advanced pathological stage, but not with tumor size, estrogen receptor (ER) and progestogen receptor (PR expression) (13). Based on the TCGA data, high level PTBP1 was found to be related with advanced T stage, AJCC stage and more metastasis in clear-cell renal cell cancer, but not with gender, age, lymph node metastasis (17). The present study revealed the clinical significance of PTBP1 based on a large cohort of gastric cancer patients, finding that high PTBP1 expression was closely correlated with gender, age, histological type, lymph node metastasis and TNM stage. These results indicated that PTBP1 was connected with and may be involved in the tumor malignant biobehaviors, especially metastasis; its upregulation was likely an important event in the neoplastic transformation and tumor progression. Expression of PTBP1 had a prognostic and predictive potential in cancers. However, in 158 patients with stages II/III patients with colorectal cancer, PTBP1 expression was proved to be not related with any of the clinicopathological parameters including gender, age, tumor size, histological type, tumor grade, tumor status, and TNM stage (14). These differences may be due in part to different tumor types and limitation of sample size. Hence, the genetic basis of these differences requires further investigation in diverse types of tumors with more patients involved in.
With regard to the biological role of PTBP1 in cancers, accumulating evidences have proved that PTBP1 contributed to tumor cell proliferation, invasion and migration in a variety of tumors including breast cancer (13), colorectal cancer (14), clear-cell renal cell carcinoma (17), lung cancer (19) and so on. In line with these previous studies, our data showed that the proliferation capacity, migratory and invasive ability were significantly suppressed in gastric cancer cells in vitro. These results demonstrated that PTBP1 had a general role in tumorigenesis and tumor progression in various type of cancers. Additionally, PTBP1 was reported to be involved in chemoradiation resistance and energy metabolism of tumor cells. For instance, PTBP1 knock-down enhanced chemoresistant osteosarcoma responsiveness to cisplatin treatment (22,23). In colon cancer, PTBP1 knockdown enhanced the sensitivity of drug-resistant cancer cells to vincristine and oxaliplatin through repression of glycolysis (15). Although previous studies uncovered that PTBP1 played a wide-ranging role in tumorigenesis and tumor progression in various cancers, intensive further researches were needed to delineate the full figure of PTBP1 in gastric cancer.
Previous studies found that PTBP1 exerted its functions in cancers predominantly through acting as an alternative splicing regulator. Its target genes contained pyruvate kinase M (PKM) (9,10,17,24,25), cell division cycle 42 (CDC42) (26), CD44V8-10 (27), myeloid ectopic viral integration site 2 (MEIS2) (24), ubiquitin-specific protease 5 (USP5) (28) etc. Among these, PKM has been mostly investigated. PTBP1 increased the transformation of PKM1 to PKM2, whose switching can reprogram metabolic pathways and promote tumorigenesis, thus promoting proliferation, invasion and migration of breast cancer (29), clear-cell renal cell carcinoma (17), and gastric cancer (21). In addition, in pancreatic cancer (25) and colon cancer (15), PTBP1 promoted drug resistance also through modulation of PKM alternative splicing. As for gastric cancer, although the effect of miR-133b/PTBP1/PKM axis on growth has been reported (21), other functions of PTBP1 and its target gene beside PKM remains to be further explored.
This study has several limitations which should be considered. First, only data from TCGA database was incorporated into bioinformatics analysis without additional external datasets. Secondary, the present study was a retrospective study in single center, prospective studies and multiple center data were needed to validate our findings. Then, the knock-in experiment was needed to further confirm the biological function of PTBP1 in gastric cancer. Besides, the molecular mechanisms underlying PTBP1 in gastric cancer tumorigenesis and development have not been defined.
Conclusions
This study demonstrates that PTBP1 is highly expressed in gastric cancer and closely related with several clinicopathological features. It could be an independent risk factor for prognosis in gastric cancer patients and act as an oncogene gene in promoting cell proliferation, invasion and metastasis of gastric cancer. Thus, PTBP1 plays an important role in gastric cancer development and progression and is expected to be a novel biomarker for targeted therapy and prediction of poor prognosis.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (Grant Nos. 81802431 and 81972516), Natural Science Foundation of Guangdong Province (Grant Nos. 2018A030313050 and 2021A1515012448) and President Foundation of Nanfang Hospital of Southern Medical University (Grant No. 2018B012).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-303/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-303/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-303/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Machlowska J, Baj J, Sitarz M, et al. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci 2020;21:4012. [Crossref] [PubMed]
- Russo AE, Strong VE. Gastric Cancer Etiology and Management in Asia and the West. Annu Rev Med 2019;70:353-67. [Crossref] [PubMed]
- Ilson DH. Advances in the treatment of gastric cancer. Curr Opin Gastroenterol 2017;33:473-6. [Crossref] [PubMed]
- Noiret M, Audic Y, Hardy S. Expression analysis of the polypyrimidine tract binding protein (PTBP1) and its paralogs PTBP2 and PTBP3 during Xenopus tropicalis embryogenesis. Int J Dev Biol 2012;56:747-53. [Crossref] [PubMed]
- La Porta J, Matus-Nicodemos R, Valentín-Acevedo A, et al. The RNA-Binding Protein, Polypyrimidine Tract-Binding Protein 1 (PTBP1) Is a Key Regulator of CD4 T Cell Activation. PLoS One 2016;11:e0158708. [Crossref] [PubMed]
- Rawcliffe DFR, Österman L, Nordin A, et al. PTBP1 acts as a dominant repressor of the aberrant tissue-specific splicing of ISCU in hereditary myopathy with lactic acidosis. Mol Genet Genomic Med 2018;6:887-97. [Crossref] [PubMed]
- Vuong JK, Lin CH, Zhang M, et al. PTBP1 and PTBP2 Serve Both Specific and Redundant Functions in Neuronal Pre-mRNA Splicing. Cell Rep 2016;17:2766-75. [Crossref] [PubMed]
- Zhang H, Wang D, Li M, et al. Metabolic and Proliferative State of Vascular Adventitial Fibroblasts in Pulmonary Hypertension Is Regulated Through a MicroRNA-124/PTBP1 (Polypyrimidine Tract Binding Protein 1)/Pyruvate Kinase Muscle Axis. Circulation 2017;136:2468-85. [Crossref] [PubMed]
- Zhu W, Zhou BL, Rong LJ, et al. Roles of PTBP1 in alternative splicing, glycolysis, and oncogensis. J Zhejiang Univ Sci B 2020;21:122-36. [Crossref] [PubMed]
- Zhong Y, Hu Z, Wu J, et al. STAU1 selectively regulates the expression of inflammatory and immune response genes and alternative splicing of the nerve growth factor receptor signaling pathway. Oncol Rep 2020;44:1863-74. [Crossref] [PubMed]
- Pina JM, Hernandez LA, Keppetipola NM. Polypyrimidine tract binding proteins PTBP1 and PTBP2 interact with distinct proteins under splicing conditions. PLoS One 2022;17:e0263287. [Crossref] [PubMed]
- Arake de Tacca LM, Pulos-Holmes MC, Floor SN, et al. PTBP1 mRNA isoforms and regulation of their translation. RNA 2019;25:1324-36. [Crossref] [PubMed]
- Wang X, Li Y, Fan Y, et al. PTBP1 promotes the growth of breast cancer cells through the PTEN/Akt pathway and autophagy. J Cell Physiol 2018;233:8930-9. [Crossref] [PubMed]
- Wang ZN, Liu D, Yin B, et al. High expression of PTBP1 promote invasion of colorectal cancer by alternative splicing of cortactin. Oncotarget 2017;8:36185-202. [Crossref] [PubMed]
- Cheng C, Xie Z, Li Y, et al. PTBP1 knockdown overcomes the resistance to vincristine and oxaliplatin in drug-resistant colon cancer cells through regulation of glycolysis. Biomed Pharmacother 2018;108:194-200. [Crossref] [PubMed]
- Jiang D, Zhang Y, Yang L, et al. Long noncoding RNA HCG22 suppresses proliferation and metastasis of bladder cancer cells by regulation of PTBP1. J Cell Physiol 2020;235:1711-22. [Crossref] [PubMed]
- Jiang J, Chen X, Liu H, et al. Polypyrimidine Tract-Binding Protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am J Cancer Res 2017;7:245-59. [PubMed]
- Kang H, Heo S, Shin JJ, et al. A miR-194/PTBP1/CCND3 axis regulates tumor growth in human hepatocellular carcinoma. J Pathol 2019;249:395-408. [Crossref] [PubMed]
- Li S, Shen L, Huang L, et al. PTBP1 enhances exon11a skipping in Mena pre-mRNA to promote migration and invasion in lung carcinoma cells. Biochim Biophys Acta Gene Regul Mech 2019;1862:858-69. [Crossref] [PubMed]
- Zhu L, Wei Q, Qi Y, et al. PTB-AS, a Novel Natural Antisense Transcript, Promotes Glioma Progression by Improving PTBP1 mRNA Stability with SND1. Mol Ther 2019;27:1621-37. [Crossref] [PubMed]
- Sugiyama T, Taniguchi K, Matsuhashi N, et al. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci 2016;107:1767-75. [Crossref] [PubMed]
- Cheng C, Ding Q, Zhang Z, et al. PTBP1 modulates osteosarcoma chemoresistance to cisplatin by regulating the expression of the copper transporter SLC31A1. J Cell Mol Med 2020;24:5274-89. [Crossref] [PubMed]
- Zhang Q, Wu J, Zhang X, et al. Transcription factor ELK1 accelerates aerobic glycolysis to enhance osteosarcoma chemoresistance through miR-134/PTBP1 signaling cascade. Aging (Albany NY) 2021;13:6804-19. [Crossref] [PubMed]
- Xie R, Chen X, Chen Z, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett 2019;449:31-44. [Crossref] [PubMed]
- Calabretta S, Bielli P, Passacantilli I, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene 2016;35:2031-9. [Crossref] [PubMed]
- He X, Yuan C, Yang J. Regulation and functional significance of CDC42 alternative splicing in ovarian cancer. Oncotarget 2015;6:29651-63. [Crossref] [PubMed]
- Takahashi H, Nishimura J, Kagawa Y, et al. Significance of Polypyrimidine Tract-Binding Protein 1 Expression in Colorectal Cancer. Mol Cancer Ther 2015;14:1705-16. [Crossref] [PubMed]
- Izaguirre DI, Zhu W, Hai T, et al. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog 2012;51:895-906. [Crossref] [PubMed]
- He X, Arslan AD, Ho TT, et al. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis 2014;3:e84. [Crossref] [PubMed]


