
Elevated expression of protein-L-isoaspartate O-methyltransferase-1 (PCMT1) in cervical cancer
Introduction
Among women, cervical cancer is the fourth most frequently diagnosed tumor worldwide, with an estimated 0.57 million cases and 0.31 million deaths according to the latest global cancer statistics (1). At present, the therapeutic strategies for cervical cancer include surgery, chemotherapy, radiotherapy and combined therapy. Although great improvements have been attained in the treatment of cervical cancer, the survival rate of patients with advanced disease remains poor (2). At present, a variety of molecular biology, genetics, targeted antitumor drugs and diagnostic, prognostic and predictive biomarkers are being sought. Ki-67, cyclin D1, p53, p63 and p16INK4a are widely used in the clinical work but insufficient in differential diagnosis and prognosis estimation (3). Finding reliable genetic, molecular and immunohistochemical markers for early diagnosis of cervical precancerous/cancerous lesions and neoplastic processes, as well as prognosis estimation remains an important task.
Post-translational modifications, such as isoaspartate in peptides and proteins, play various roles in the development of human disease. The protein-L-isoaspartate O-methyltransferase-1 (PCMT1), a member of the type II class of protein carboxyl methyltransferase enzymes, has been found to be involved in the repair of intracellular protein in multiple tissues via recognizing and converting L-isoaspartyl and D-aspartyl (4). Accumulating evidence indicates that PCMT1 is increased in brain tissue and is associated with Parkinson’s disease (5,6). Pyun et al. (7) reported that four polymorphisms in PCMT1 were also correlated with premature ovarian failure (POF). In human malignancies, PCMT1 was upregulated in bladder cancer tissues and function as an independent unfavorable prognostic factor for patients with bladder cancer (8). However, the expression of PCMT1 in cervical cancer is still unclear.
In current study, we focus on the alterations in the expression of PCMT1 in cervical cancer. Immunohistochemistry (IHC) and public databases analysis were used to show the expression pattern of PCTM1 in different pathological types of cervical tissues. The prognostic value in cervical cancer were also explored. To further understand the role of PCMT1 in cervical carcinogenesis, the potential related regulatory signaling pathways were explored. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2700/rc).
Methods
Bioinformatic analysis
We used the Gene Expression Omnibus (GEO) data, which are available in a public repository from the GEO website (https://www.ncbi.nlm.nih.gov/geo/), to evaluate the expression of PCMT1 in different cervical tissues. GSE7410 includes 35 patients with or without unfavorable prognostic factors of cervical cancer patients and 5 non-cervical cancer patients who underwent hysterectomy for benign reasons. GSE6791 was used to analyze the expression of PCMT1 between 8 normal cervical epitheliums and 20 cervical cancer tissues. The Kaplan-Meier plotter (https://kmplot.com/analysis/) was used to perform survival analyses of cervical cancer patients (9). Gene set enrichment analysis (GSEA) was used to evaluate various cancer-related pathways and biological process in transcriptome levels between high and low expression of PCMT1 in cervical cancer by GSEA v3.0 software (10,11). Gene Expression Profiling Interactive Analysis 2 (GEPIA2) platform (http://gepia2.cancer-pku.cn/#index) was used to analyze the correlation between the expression of PCMT1 and carcinogenic signaling related genes (12).
Immunohistochemical analysis
The commercial human cervical tissue microarrays were purchased from Shanghai Qizhe Biotechnology (Shanghai, China). IHC staining for PCMT1 was performed according to standard protocols. Briefly, antigen retrieval was achieved by microwave using sodium citrate solution in 95 ℃ for 10 min. The microarrays were incubated with PCMT1 polyclonal antibody (10519-1-AP, diluted to 1:200; Proteintech, China) at 4 ℃ overnight. Then the microarrays were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h. All IHC microarrays were reviewed independently by two investigators. We scored PCMT1 expression as immunoreactivity score (IRS) according to the following criteria. The staining intensity (0, negative; 1, weak; 2, moderate; and 3, strong) and the staining intensity (0, no positive cells; 1, <10%; 2, 10–33%; 3, 34–66%; and 4, ≥67%) that expressed PCMT1 were evaluated for each case. The IRS value was calculated by multiplying the staining extent by the staining intensity.
Statistical analysis
The Student’s t-test was used to compare the gene expression between different groups. Kaplan-Meier analysis was used to evaluate the survival rate of cervical cancer patients. Statistical analyses were performed in GraphPad Prism (version 8.0, GraphPad Software, La Jolla, CA, USA). A P value <0.05 (two-sided) indicated a statistically significant difference.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The current study was approved by the Institutional Ethics Committee of Shanghai General Hospital (No. AF09). The participants gave informed consent before taking part.
Results
The expression of PCMT1 in cervical tissues
To study the expression of PCMT1 in different pathological types of cervical tissues, we firstly examined the protein level of PCMT1 in normal cervix, cervical squamous intraepithelial lesion and cervical cancer tissues. As shown in Figure 1A, PCMT1-positive cells are found in almost all cervical cancer tissues and full thickness of the squamous epithelium of high-grade squamous intraepithelial lesion (HSIL). Cervical cancer (10.70±0.54) and HSIL (7.40±0.42) had higher IRS of PCMT1 compared to normal cervix (5.00±0.86) and low-grade squamous intraepithelial lesion (LSIL) (6.22±0.57) (P<0.05; Figure 1A,1B). Moreover, the positive staining of PCMT1 was increased with the rise of the grade of intraepithelial lesion (P<0.05), but there was no significant difference between normal cervix and LSIL (P>0.05; Figure 1A,1B).
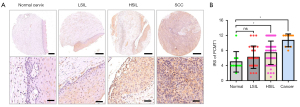
The expression of PCMT1 in cervical cancer and adjacent non-cancerous tissues
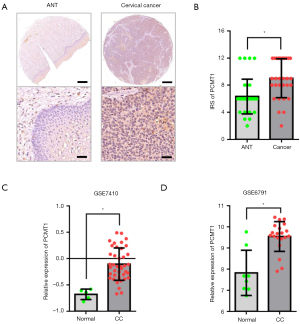
In order to indicate the potential role of PCMT1 in cervical cancer, we next investigated the protein level of PCMT1 in cervical cancer and paired adjacent non-cancerous tissues. Consistent with the expression pattern of PCMT1 mentioned above, the IRS of PCMT1 in cervical cancer tissues was significantly higher than that in paired adjacent non-cancerous tissues (9.03±0.52 vs. 6.32±0.46, P<0.05; Figure 2A,2B). Additionally, we confirmed the above findings by analyzing the GEO databases. As demonstrated in Figure 2C,2D, the mRNA expression of PCMT1 was upregulated in cervical cancer compared to normal cervix tissues (P<0.05; GSE7410, GSE6791).
The expression of PCMT1 predicted worse survival of cervical cancer patients
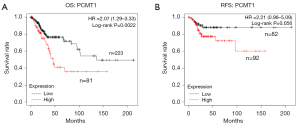
To investigate the relationship between the expression of PCMT1 and the survival rate in cervical cancer patients, we studied the prognosis of cervical cancer patients from the Kaplan-Meier plotter database. The expression of PCMT1 was highly associated with the overall survival (OS) of cervical cancer patients (P=0.0022; log-rank test). The PCMT1-high group was correlated with worse survival in cervical cancer, whereas the PCMT1-low group had a better survival (Figure 3A). Moreover, the relapse-free survival (RFS) was also analyzed. As shown in Figure 3B, high expression of PCMT1 correlated with poor RFS, although there was no statistical difference (P=0.056; log-rank test).
The relationship between PCMT1 and oncogenic pathways in cervical cancer
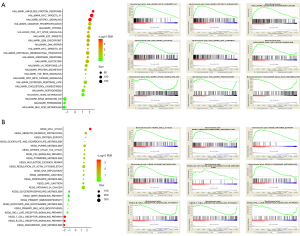
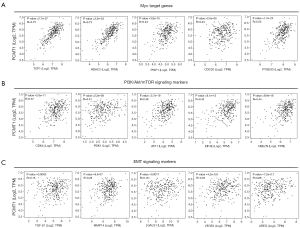
To reveal the potential role of PCMT1 in cervical cancer, hallmark-related pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis are performed by using GSEA. We demonstrated that PCMT1 was involved in the unfolded protein response, Myc targets, mTORC1 signaling, oxidative phosphorylation, hypoxia, PI3K/Akt signaling, E2F targets, DNA repair, epithelial mesenchymal transition (EMT), etc. (Figure 4A). Furthermore, KEGG pathway enrichment showed the potential biological processes regulated by PCMT1. The expression of PCMT1 was positively enriched in cell cycle, ubiquitin mediated proteolysis, protein export, citrate cycle TCA cycle, p53 signaling pathway and DNA replication, whereas arachidonic acid metabolism, T cell and B cell receptor signaling pathway, primary immunodeficiency and glycerophospholipid metabolism were negatively associated with the expression of PCMT1 (Figure 4B). Additionally, correlation analysis also showed that the expression of PCMT1 was significantly correlated with Myc target genes (including TCP1, HDAC1, PWP1, CDC20 and PTGES3; Figure 5A), PI3K/Akt/mTOR signaling (including CDK4, PDK1, ATF1, EIF4E and UBE2N; Figure 5B) and EMT signaling (including TGF-β1, MMP14, LGALS1, VEGFA and AREG; Figure 5C).
Discussion
In this study, we explored the expression pattern, prognostic value and potential function of PCMT1 in cervical cancer. We validated the protein level of PCMT1 in different pathological types of cervical tissues via IHC assays and analyzed relationships between its expression with survival rate of cervical cancer patient. GSEA had also been performed to show the signaling pathways potentially regulated by PCMT1.
Protein methyltransferases (PMTs) are classified into three classes according to their preferred amino acids (arginine, glutamine/aspartate, or lysine) for methylation (13). The biological roles of PMTs in mammalian cells have been widely studied, including aging, protein repair, cell response to stress, and carcinogenesis, etc. (14). In multiple human solid tumors, including cervical cancer, the histone methyltransferase NSD2 is frequently over-expressed (15). The protein lysine methyltransferase SETD8 has been found to regulate cancer cell proliferation (16), metabolism (17), and stemness characteristics (18). The histone methyltransferase SMYD2 facilitates cancer cell malignancy and drug resistance (19-21). The protein arginine methyltransferase PRMT5 not only controls oncogenic processes, but also mediates tumor microenvironmental signaling pathways (22). As such, targeting PMTs has emerged as a potential therapeutic strategy with several chemical inhibitors and antagonists (14).
Protein iso-aspartate methyltransferases (PIMTs) that convert iso-aspartate to aspartate in proteins have been identified as important factors in Alzheimer’s disease and cancers (23). Strong PIMT expression was particularly observed in advanced stages of lung cancer, and was associated with a shorter survival rate (24). Lee et al. (25) reported that PIMT reduces the protein level of tumor suppressor protein p53 through carboxyl methylation, and suppresses the p53-mediated transcription of target genes in cancer cell lines. PCMT1 is one of the PIMT variant in human, and has been found over-expressed in bladder cancer (8). However, the expression alteration of PCMT1 in cervical cancer is still unclear. In this research, we showed for the first time that PCMT1 was overexpressed in cervical high-grade intra-epithelial neoplasia and cervical cancer by using IHC and bioinformatic analysis. Moreover, high expression of PCMT1 was also correlated with worse OS rate of cervical cancer patients, which indicated that PCMT1 might be a potential predictive biomarker for cervical cancer.
According to GSEA, high expression of PCMT1 in cervical cancer was associated to a few gene sets that are closely related to carcinogenesis, including Myc targets, PI3K/Akt/mTOR signaling, E2F targets, DNA repair and EMT. Previous studies have confirmed that dysregulation of Myc targets, PI3K/Akt/mTOR signaling and EMT signaling could mediate the development and progression of cervical cancer (26-28). We found that PCMT1 expression was significantly associated with Myc target genes (such as TCP1, HDAC2 and PWP1), PI3K/Akt/mTOR signaling markers (such as ATF1, PDK1 and EIF4) and EMT signaling markers (such as TGF-β1, LAGLS1 and VEGFA), suggesting that these carcinogenic signaling could be further studied to understand the mechanism of PCMT1 in cervical cancer.
Although we have revealed the potential role of PCMT1 in cervical cancer, there is still some limitations which could be future studied. Firstly, while clinical evidence indicated that PCMT1 might have influence on cervical carcinogenesis, we have not provided functional verification to confirm the role of PCMT1 in vivo or in vitro, which might be different from its expression pattern. Secondly, although GSEA demonstrated that PCMT1 was related to multiple signaling pathways, more mechanistic studies need to confirm these findings.
In summary, our present study showed that the expression of PCMT1, both in the mRNA and protein level, was increased in cervical cancer tissues compared with that in normal cervix and adjacent non-cancerous tissues. Moreover, PCMT1 expression was significantly correlated with the OS of cervical cancer patients and a variety of distinct oncogenic pathways. Therefore, PCMT1 might serve as a biomarker and a potential therapeutic target for cervical cancer.
Acknowledgments
Funding: This study was supported financially by the National Natural Science Foundation of China (Grant No. 82072868).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2700/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2700/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2700/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The current study was approved by the Institutional Ethics Committee of Shanghai General Hospital (No. AF09). The participants gave informed consent before taking part.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Shrestha AD, Neupane D, Vedsted P, et al. Cervical Cancer Prevalence, Incidence and Mortality in Low and Middle Income Countries: A Systematic Review Asian Pac J Cancer Prev 2018;19:319-24. [PubMed]
- Volkova LV, Pashov AI, Omelchuk NN. Cervical Carcinoma: Oncobiology and Biomarkers. Int J Mol Sci 2021;22:12571. [Crossref] [PubMed]
- DeVry CG, Clarke S. Polymorphic forms of the protein L-isoaspartate (D-aspartate) O-methyltransferase involved in the repair of age-damaged proteins. J Hum Genet 1999;44:275-88. [Crossref] [PubMed]
- Lowenson JD, Kim E, Young SG, et al. Limited accumulation of damaged proteins in l-isoaspartyl (D-aspartyl) O-methyltransferase-deficient mice. J Biol Chem 2001;276:20695-702. [Crossref] [PubMed]
- Liscovitch N, French L. Differential Co-Expression between α-Synuclein and IFN-γ Signaling Genes across Development and in Parkinson's Disease. PLoS One 2014;9:e115029. [Crossref] [PubMed]
- Pyun JA, Kang H, Lee SK, et al. Association between polymorphisms in the protein L-isoaspartate (D-aspartate) O-methyltransferase gene and premature ovarian failure. Fertil Steril 2009;91:1362-5. [Crossref] [PubMed]
- Dong L, Li Y, Xue D, et al. PCMT1 is an unfavorable predictor and functions as an oncogene in bladder cancer. IUBMB Life 2018;70:291-9. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Clarke SG. Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem Sci 2013;38:243-52. [Crossref] [PubMed]
- Dilworth D, Barsyte-Lovejoy D. Targeting protein methylation: from chemical tools to precision medicines. Cell Mol Life Sci 2019;76:2967-85. [Crossref] [PubMed]
- Chen R, Chen Y, Zhao W, et al. The Role of Methyltransferase NSD2 as a Potential Oncogene in Human Solid Tumors. Onco Targets Ther 2020;13:6837-46. [Crossref] [PubMed]
- Liu M, Qin Y, Hu Q, et al. SETD8 potentiates constitutive ERK1/2 activation via epigenetically silencing DUSP10 expression in pancreatic cancer. Cancer Lett 2021;499:265-78. [Crossref] [PubMed]
- Huang R, Yu Y, Zong X, et al. Monomethyltransferase SETD8 regulates breast cancer metabolism via stabilizing hypoxia-inducible factor 1α. Cancer Lett 2017;390:1-10. [Crossref] [PubMed]
- Piao L, Che N, Li H, et al. SETD8 promotes stemness characteristics and is a potential prognostic biomarker of gastric adenocarcinoma. Exp Mol Pathol 2020;117:104560. [Crossref] [PubMed]
- Komatsu S, Imoto I, Tsuda H, et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 2009;30:1139-46. [Crossref] [PubMed]
- Komatsu S, Ichikawa D, Hirajima S, et al. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer 2015;112:357-64. [Crossref] [PubMed]
- Ren H, Wang Z, Chen Y, et al. SMYD2-OE promotes oxaliplatin resistance in colon cancer through MDR1/P-glycoprotein via MEK/ERK/AP1 pathway. Onco Targets Ther 2019;12:2585-94. [Crossref] [PubMed]
- Kim H, Ronai ZA. PRMT5 function and targeting in cancer. Cell Stress 2020;4:199-215. [Crossref] [PubMed]
- Mishra PKK, Mahawar M. PIMT-Mediated Protein Repair: Mechanism and Implications. Biochemistry (Mosc) 2019;84:453-63. [Crossref] [PubMed]
- Saito H, Yamashita M, Ogasawara M, et al. Chaperone protein L-isoaspartate (D-aspartyl) O-methyltransferase as a novel predictor of poor prognosis in lung adenocarcinoma. Hum Pathol 2016;50:1-10. [Crossref] [PubMed]
- Lee JC, Kang SU, Jeon Y, et al. Protein L-isoaspartyl methyltransferase regulates p53 activity. Nat Commun 2012;3:927. [Crossref] [PubMed]
- Higareda-Almaraz JC, Enríquez-Gasca Mdel R, Hernández-Ortiz M, et al. Proteomic patterns of cervical cancer cell lines, a network perspective. BMC Syst Biol 2011;5:96. [Crossref] [PubMed]
- Bossler F, Hoppe-Seyler K, Hoppe-Seyler F. PI3K/AKT/mTOR Signaling Regulates the Virus/Host Cell Crosstalk in HPV-Positive Cervical Cancer Cells. Int J Mol Sci 2019;20:2188. [Crossref] [PubMed]
- Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett 2015;356:321-31. [Crossref] [PubMed]


