
A network pharmacology-based exploration of the active compounds and potential drug targets of Si-Jun-Zi decoction in the treatment of cutaneous squamous cell carcinoma
Introduction
Cutaneous squamous cell carcinoma (cSCC) is a kind of skin tumor with an incidence second only to that of basal cell carcinoma (1). It arises from keratinocytes in the epidermis or appendages. Currently, management of cSCC involves surgical treatment, radiation therapy, chemotherapy, and targeted therapy. Surgical treatment is the main method for early cSCC. However, the survival problems faced by cSCC patients include postoperative recurrence, lymph node metastasis, and distant metastasis, all of which require clinical solutions (2,3). Although targeted therapy is the main treatment for these cSCC patients, the prognosis remains poor (4).
Network pharmacology is a new research method combining pharmacology and pharmacodynamics based on a variety of network databases, which can help us understand the interactions among traditional Chinese medicines (TCM), compounds, disease, and targets in a more systematic and comprehensive way.
For a long period of time, TCM has successfully treated a variety of complex diseases through the use of multi-compound decoctions aimed at multiple targets. Through online pharmacology, researchers can not only explore the compounds of TCM formulae, but also understand the interaction between active ingredients and their related targets, which provides a new, highly applicable method for clarifying the mechanism of TCM treatment of diseases.
Si Jun Zi (SJZ) decoction is a classical Chinese medicine formula extracted from Ginseng (Ren Shen), Glycyrrhiza inflata (Gan Cao), Largehead Atractylodes (Bai Zhu), and Indian Bread (Fu Ling) in a ratio of 3:3:3:2. It has been used as a main therapeutic method or complementary therapy for the treatment of various diseases, including cancer, for more than 1,000 years. Given that the function of inhibiting tumor growth, improving tumor cachexia, regulating tumor microenvironment, and regulating body immunity, the SJZ decoction is widely used to facilitate the swift recovery of cancer patients, since disease or chemotherapy may lead to poor physical fitness (5). Nowadays, the research of SJZ Decoction has been reported internationally for the treatment of esophageal squamous cell carcinoma and lung squamous cell carcinoma, but there are no reports or studies on the treatment of cSCC. The main compounds and mechanisms of SJZ decoction in the treatment of tumors have not been clarified. As the main active constituent in ginseng, ginsenosides have been widely used in the treatment of photoaging, hair loss, and trauma, among others (6). In recent years, Licochalcone A, Licochalcone B, and Licochalcone D have been reported to inhibit the growth of lung cancer cells through different pathways (7-11). Licochalcone C, Licochalcone H, and GIP1 can induce apoptosis of human oral squamous cell carcinoma cells (HOSCC) (12). Studies have found that Atractylodes I is the main effective ingredient in alleviating the symptoms of gastric cancer, and it can also improve the occurrence and development of melanoma to a certain extent (13,14). Currently, the main active ingredients of SJZ decoction, which is made up of four Chinese herbs, remain to be confirmed.
Therefore, in this study, we used network pharmacology and bioinformatics analysis to try to construct the action network of SJZ decoction and cSCC, with the aim of solving the following questions: (I) what are the active ingredients of SJZ decoction; (II) which cSCC disease targets are related to the active compounds of SJZ decoction; and (III) what new information can network pharmacology yield in the study of the treatment of cSCC by SJZ Decoction.
Methods
Active ingredients and potential targets of TCMs
We used three databases to identify the chemical ingredients in Ginseng, Glycyrrhiza inflata, Largehead Atractylodes, and Indian Bread: (I) Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP; available online: https://old.tcmsp-e.com/tcmsp.php); (II) Traditional Chinese Medicines Integrated Database (TCMID; available online: http://47.100.169.139/tcmid/search/); and (III) TCM Database@Taiwan (http://tcm.cmu.edu.tw/). Specified oral bioavailability (OB) and drug-likeness (DL) properties were treated as filter criteria in the TCMSPTM, which may allow the various components of the drug to perform the desired activity. Compounds that met both criteria were considered as candidate molecules.
Network analysis and building
Constructing a ‘drug-target-disease-pathway’ network allows us to gather a variety of information regarding active ingredients and their corresponding targets and to better understand the anti-tumor mechanism of active ingredients from another perspective. Firstly, we sorted out the screened active ingredients and corresponding potential targets of the four TCM ingredients. Then, we extracted cSCC disease-related targets from GeneCards online database (https://www.genecards.org/) for sorting and conducted functional enrichment analysis of these common potential targets, including “Gene Ontology (GO)” and “Kyoto Encyclopedia of Genes and Genomes (KEGG)“ analyses. Finally, all data were processed by Cytoscape 3.5.1 software (https://cytoscape.org/) to construct a complex network of “drug-component-target-pathway-disease”.
Plant sample preparation
The SJZ decoction is a mixture of Ginseng, Indian Bread, Largehead Atractylodes, and Glycyrrhiza inflata at a mass ratio of 3:3:3:2. The components were purchased from the Chinese Pharmacy of the Guangdong Second Provincial General Hospital (GD2H; Guangzhou, China). A total of 1,650 g of the Chinese herbal medicine (CHM) formula consisting of Ginseng, Indian Bread, Largehead Atractylodes, and Glycyrrhiza inflata was soaked for 30 minutes and extracted with 100 ℃ water twice. The sample was then cooled and concentrated.
Ultra-high performance liquid chromatography for sample
An ultra-high performance liquid chromatography (UPLC) system (Water, Milford, MA, USA) was used for dealing with the filtered sample solutions, combining with the annotation and classification of mass spectrometry database information were completed using precise characterization Instrument ASTAT-DAP. LC-MS (Thermo, Ultimate 3000 LC, HF) fitted with a C18 column [Zorbax Eclipse C18 (1.8 µm × 2.1 mm × 100 mm)] and the separation conditions were as follows: column temperature =30 ℃; flow rate =0.3 mL/min; mobile phase A=water + 0.1% formic acid, mobile phase B = pure acetonitrile; Injection volume =2 µ; and active autokinetic nozzle =4 ℃.
Statistical analysis
In this study, TCMSP, TCMID, TCM Database@Taiwan, and GeneCards database were used for data acquisition, David online tool and Cytoscape software were used for data analysis and image rendering respectively.
Results
cSCC-related gene pathways and networks
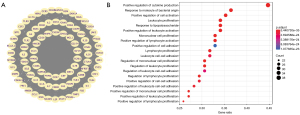
A total of 748 human genes with a high score (≥30.0) associated with cSCC were identified in the GeneCard database, and the encoded proteins were assembled into a set of 94 pathways and 25 networks using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; (https://cn.string-db.org/cgi/input.pl) and Cytoscape software. Cluster 1, which has the highest relevant scores among the networks, is shown in Figure 1A. The top five pathways involved cytokine-cytokine receptor interaction, lipid and atherosclerosis, rheumatoid, arthritis, malaria, and hepatitis B. The GO enrichment and network analysis showed that “T positive regulation of cytokine production”, “response to molecule of bacterial origin”, “leukocyte proliferation, positive regulation of mononuclear cell proliferation”, and “regulation of mononuclear cell proliferation” covered the top 5 biological processes of cSCC target proteins (Figure 1B).
Screening of common targets of TCM compound and cSCC
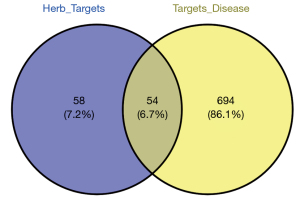
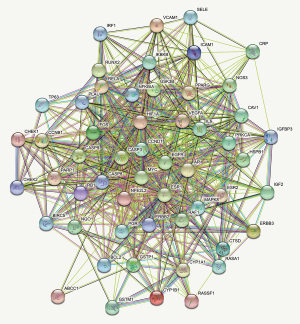
The targets of the four herbs were predicted using the TCMSP database. A total of 136 candidate compounds were screened from 559 chemicals with DL (≥0.18) and OB (≥30%), including 22 in Ginseng, 7 in Largehead Atractylodes, 15 in Indian Bread, and 92 in Glycyrrhiza inflata. Several compounds have been reported to have more than one biological activity in recent studies. A total of 112 different herb targets obtained from 136 candidate compounds (Table 1) were further subjected to the UniProt (https://www.uniprot.org/) for conversing ID name. A total of 54 common candidate targets (Table 2) were selected from 748 different candidate cSCC targets filtered from GeneCards and 112 herb targets (Figure 2). The STRING database was used to construct the protein-protein interaction (PPI) networks (Figure 3).
Table 1
| Resource | Mol ID | Molecule name | OB (%) | DL |
|---|---|---|---|---|
| Ginseng | MOL005399 | alexandrin_qt | 36.91 | 0.75 |
| Ginseng | MOL005308 | Aposiopolamine | 66.65 | 0.22 |
| Ginseng | MOL005320 | arachidonate | 45.57 | 0.2 |
| Ginseng | MOL000358 | beta-sitosterol | 36.91 | 0.75 |
| Ginseng | MOL005314 | Celabenzine | 101.88 | 0.49 |
| Ginseng | MOL004492 | Chrysanthemaxanthin | 38.72 | 0.58 |
| Ginseng | MOL005317 | Deoxyharringtonine | 39.27 | 0.81 |
| Ginseng | MOL005318 | Dianthramine | 40.45 | 0.2 |
| Ginseng | MOL002879 | Diop | 43.59 | 0.39 |
| Ginseng | MOL005321 | Frutinone A | 65.9 | 0.34 |
| Ginseng | MOL000787 | Fumarine | 59.26 | 0.83 |
| Ginseng | MOL005401 | ginsenoside Rg5_qt | 39.56 | 0.79 |
| Ginseng | MOL005344 | ginsenoside rh2 | 36.32 | 0.56 |
| Ginseng | MOL005348 | Ginsenoside-Rh4_qt | 31.11 | 0.78 |
| Ginseng | MOL005356 | Girinimbin | 61.2 | 0.31 |
| Ginseng | MOL005357 | Gomisin B | 31.99 | 0.83 |
| Ginseng | MOL003648 | Inermin | 65.83 | 0.54 |
| Ginseng | MOL000422 | kaempferol | 41.88 | 0.24 |
| Ginseng | MOL005360 | malkangunin | 57.71 | 0.63 |
| Ginseng | MOL005376 | Panaxadiol | 33.09 | 0.79 |
| Ginseng | MOL000449 | Stigmasterol | 43.83 | 0.76 |
| Ginseng | MOL005384 | suchilactone | 57.52 | 0.56 |
| Largehead Atractylodes | MOL000020 | 12-senecioyl-2E,8E,10E-atractylentriol | 62.4 | 0.22 |
| Largehead Atractylodes | MOL000021 | 14-acetyl-12-senecioyl-2E,8E,10E-atractylentriol | 60.31 | 0.31 |
| Largehead Atractylodes | MOL000022 | 14-acetyl-12-senecioyl-2E,8Z,10E-atractylentriol | 63.37 | 0.3 |
| Largehead Atractylodes | MOL000028 | α-Amyrin | 39.51 | 0.76 |
| Largehead Atractylodes | MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | 36.23 | 0.78 |
| Largehead Atractylodes | MOL000049 | 3β-acetoxyatractylone | 54.07 | 0.22 |
| Largehead Atractylodes | MOL000072 | 8β-ethoxy atractylenolide III | 35.95 | 0.21 |
| Indian Bread | MOL000273 | (2R)-2-[(3S,5R,10S,13R,14R,16R,17R)-3,16-dihydroxy-4,4,10,13,14-pentamethyl-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta[a]phenanthren-17-yl]-6-methylhept-5-enoic acid | 30.93 | 0.81 |
| Indian Bread | MOL000275 | Trametenolic acid | 38.71 | 0.8 |
| Indian Bread | MOL000276 | 7,9(11)-dehydropachymic acid | 35.11 | 0.81 |
| Indian Bread | MOL000279 | Cerevisterol | 37.96 | 0.77 |
| Indian Bread | MOL000280 | (2R)-2-[(3S,5R,10S,13R,14R,16R,17R)-3,16-dihydroxy-4,4,10,13,14-pentamethyl-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta[a]phenanthren-17-yl]-5-isopropyl-hex-5-enoic acid | 31.07 | 0.82 |
| Indian Bread | MOL000282 | ergosta-7,22E-dien-3beta-ol | 43.51 | 0.72 |
| Indian Bread | MOL000283 | Ergosterol peroxide | 40.36 | 0.81 |
| Indian Bread | MOL000285 | (2R)-2-[(5R,10S,13R,14R,16R,17R)-16-hydroxy-3-keto-4,4,10,13,14-pentamethyl-1,2,5,6,12,15,16,17-octahydrocyclopenta[a]phenanthren-17-yl]-5-isopropyl-hex-5-enoic acid | 38.26 | 0.82 |
| Indian Bread | MOL000287 | 3beta-Hydroxy-24-methylene-8-lanostene-21-oic acid | 38.7 | 0.81 |
| Indian Bread | MOL000289 | Pachymic acid | 33.63 | 0.81 |
| Indian Bread | MOL000290 | Poricoic acid A | 30.61 | 0.76 |
| Indian Bread | MOL000291 | Poricoic acid B | 30.52 | 0.75 |
| Indian Bread | MOL000292 | Poricoic acid C | 38.15 | 0.75 |
| Indian Bread | MOL000296 | Hederagenin | 36.91 | 0.75 |
| Indian Bread | MOL000300 | Dehydroeburicoic acid | 44.17 | 0.83 |
| Glycyrrhiza inflata | MOL000098 | Quercetin | 46.43 | 0.28 |
| Glycyrrhiza inflata | MOL000211 | Mairin | 55.38 | 0.78 |
| Glycyrrhiza inflata | MOL000239 | Jaranol | 50.83 | 0.29 |
| Glycyrrhiza inflata | MOL000354 | Isorhamnetin | 49.6 | 0.31 |
| Glycyrrhiza inflata | MOL000359 | Sitosterol | 36.91 | 0.75 |
| Glycyrrhiza inflata | MOL000392 | Formononetin | 69.67 | 0.21 |
| Glycyrrhiza inflata | MOL000417 | Calycosin | 47.75 | 0.24 |
| Glycyrrhiza inflata | MOL000422 | Kaempferol | 41.88 | 0.24 |
| Glycyrrhiza inflata | MOL000497 | licochalcone a | 40.79 | 0.29 |
| Glycyrrhiza inflata | MOL000500 | Vestitol | 74.66 | 0.21 |
| Glycyrrhiza inflata | MOL001484 | Inermine | 75.18 | 0.54 |
| Glycyrrhiza inflata | MOL001792 | DFV | 32.76 | 0.18 |
| Glycyrrhiza inflata | MOL002311 | Glycyrol | 90.78 | 0.67 |
| Glycyrrhiza inflata | MOL002565 | Medicarpin | 49.22 | 0.34 |
| Glycyrrhiza inflata | MOL003656 | Lupiwighteone | 51.64 | 0.37 |
| Glycyrrhiza inflata | MOL003896 | 7-Methoxy-2-methyl isoflavone | 42.56 | 0.2 |
| Glycyrrhiza inflata | MOL004328 | naringenin | 59.29 | 0.21 |
| Glycyrrhiza inflata | MOL004805 | (2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one | 31.79 | 0.72 |
| Glycyrrhiza inflata | MOL004806 | Euchrenone | 30.29 | 0.57 |
| Glycyrrhiza inflata | MOL004808 | Glyasperin B | 65.22 | 0.44 |
| Glycyrrhiza inflata | MOL004810 | Glyasperin F | 75.84 | 0.54 |
| Glycyrrhiza inflata | MOL004811 | Glyasperin C | 45.56 | 0.4 |
| Glycyrrhiza inflata | MOL004814 | Isotrifoliol | 31.94 | 0.42 |
| Glycyrrhiza inflata | MOL004815 | (E)-1-(2,4-dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one | 39.62 | 0.35 |
| Glycyrrhiza inflata | MOL004820 | kanzonols W | 50.48 | 0.52 |
| Glycyrrhiza inflata | MOL004824 | (2S)-6-(2,4-dihydroxyphenyl)-2-(2-hydroxypropan-2-yl)-4-methoxy-2,3-dihydrofuro[3,2-g]chromen-7-one | 60.25 | 0.63 |
| Glycyrrhiza inflata | MOL004827 | Semilicoisoflavone B | 48.78 | 0.55 |
| Glycyrrhiza inflata | MOL004828 | Glepidotin A | 44.72 | 0.35 |
| Glycyrrhiza inflata | MOL004829 | Glepidotin B | 64.46 | 0.34 |
| Glycyrrhiza inflata | MOL004833 | Phaseolinisoflavan | 32.01 | 0.45 |
| Glycyrrhiza inflata | MOL004835 | Glypallichalcone | 61.6 | 0.19 |
| Glycyrrhiza inflata | MOL004838 | 8-(6-hydroxy-2-benzofuranyl)-2,2-dimethyl-5-chromenol | 58.44 | 0.38 |
| Glycyrrhiza inflata | MOL004841 | Licochalcone B | 76.76 | 0.19 |
| Glycyrrhiza inflata | MOL004848 | Licochalcone G | 49.25 | 0.32 |
| Glycyrrhiza inflata | MOL004849 | 3-(2,4-dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | 59.62 | 0.43 |
| Glycyrrhiza inflata | MOL004855 | Licoricone | 63.58 | 0.47 |
| Glycyrrhiza inflata | MOL004856 | Gancaonin A | 51.08 | 0.4 |
| Glycyrrhiza inflata | MOL004857 | Gancaonin B | 48.79 | 0.45 |
| Glycyrrhiza inflata | MOL004860 | Licorice glycoside E | 32.89 | 0.27 |
| Glycyrrhiza inflata | MOL004863 | 3-(3,4-dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone | 66.37 | 0.41 |
| Glycyrrhiza inflata | MOL004864 | 5,7-dihydroxy-3-(4-methoxyphenyl)-8-(3-methylbut-2-enyl)chromone | 30.49 | 0.41 |
| Glycyrrhiza inflata | MOL004866 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone | 44.15 | 0.41 |
| Glycyrrhiza inflata | MOL004879 | Glycyrin | 52.61 | 0.47 |
| Glycyrrhiza inflata | MOL004882 | Licocoumarone | 33.21 | 0.36 |
| Glycyrrhiza inflata | MOL004883 | Licoisoflavone | 41.61 | 0.42 |
| Glycyrrhiza inflata | MOL004884 | Licoisoflavone B | 38.93 | 0.55 |
| Glycyrrhiza inflata | MOL004885 | Licoisoflavanone | 52.47 | 0.54 |
| Glycyrrhiza inflata | MOL004891 | Shinpterocarpin | 80.3 | 0.73 |
| Glycyrrhiza inflata | MOL004898 | (E)-3-[3,4-dihydroxy-5-(3-methylbut-2-enyl)phenyl]-1-(2,4-dihydroxyphenyl)prop-2-en-1-one | 46.27 | 0.31 |
| Glycyrrhiza inflata | MOL004903 | Liquiritin | 65.69 | 0.74 |
| Glycyrrhiza inflata | MOL004904 | Licopyranocoumarin | 80.36 | 0.65 |
| Glycyrrhiza inflata | MOL004905 | 3,22-Dihydroxy-11-oxo-delta(12)-oleanene-27-alpha-methoxycarbonyl-29-oic acid | 34.32 | 0.55 |
| Glycyrrhiza inflata | MOL004907 | Glyzaglabrin | 61.07 | 0.35 |
| Glycyrrhiza inflata | MOL004908 | Glabridin | 53.25 | 0.47 |
| Glycyrrhiza inflata | MOL004910 | Glabranin | 52.9 | 0.31 |
| Glycyrrhiza inflata | MOL004911 | Glabrene | 46.27 | 0.44 |
| Glycyrrhiza inflata | MOL004912 | Glabrone | 52.51 | 0.5 |
| Glycyrrhiza inflata | MOL004913 | 1,3-dihydroxy-9-methoxy-6-benzofurano[3,2-c]chromenone | 48.14 | 0.43 |
| Glycyrrhiza inflata | MOL004914 | 1,3-dihydroxy-8,9-dimethoxy-6-benzofurano[3,2-c]chromenone | 62.9 | 0.53 |
| Glycyrrhiza inflata | MOL004915 | Eurycarpin A | 43.28 | 0.37 |
| Glycyrrhiza inflata | MOL004917 | Glycyroside | 37.25 | 0.79 |
| Glycyrrhiza inflata | MOL004924 | (-)-Medicocarpin | 40.99 | 0.95 |
| Glycyrrhiza inflata | MOL004935 | Sigmoidin-B | 34.88 | 0.41 |
| Glycyrrhiza inflata | MOL004941 | (2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | 71.12 | 0.18 |
| Glycyrrhiza inflata | MOL004945 | (2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | 36.57 | 0.32 |
| Glycyrrhiza inflata | MOL004948 | Isoglycyrol | 44.7 | 0.84 |
| Glycyrrhiza inflata | MOL004949 | Isolicoflavonol | 45.17 | 0.42 |
| Glycyrrhiza inflata | MOL004957 | HMO | 38.37 | 0.21 |
| Glycyrrhiza inflata | MOL004959 | 1-Methoxyphaseollidin | 69.98 | 0.64 |
| Glycyrrhiza inflata | MOL004961 | Quercetin der. | 46.45 | 0.33 |
| Glycyrrhiza inflata | MOL004966 | 3'-Hydroxy-4'-O-Methylglabridin | 43.71 | 0.57 |
| Glycyrrhiza inflata | MOL004974 | 3'-Methoxyglabridin | 46.16 | 0.57 |
| Glycyrrhiza inflata | MOL004978 | 2-[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano[6,5-f]chromen-3-yl]-5-methoxyphenol | 36.21 | 0.52 |
| Glycyrrhiza inflata | MOL004980 | Inflacoumarin A | 39.71 | 0.33 |
| Glycyrrhiza inflata | MOL004985 | icos-5-enoic acid | 30.7 | 0.2 |
| Glycyrrhiza inflata | MOL004988 | Kanzonol F | 32.47 | 0.89 |
| Glycyrrhiza inflata | MOL004989 | 6-prenylated eriodictyol | 39.22 | 0.41 |
| Glycyrrhiza inflata | MOL004990 | 7,2',4'-trihydroxy-5-methoxy-3-arylcoumarin | 83.71 | 0.27 |
| Glycyrrhiza inflata | MOL004991 | 7-Acetoxy-2-methylisoflavone | 38.92 | 0.26 |
| Glycyrrhiza inflata | MOL004993 | 8-prenylated eriodictyol | 53.79 | 0.4 |
| Glycyrrhiza inflata | MOL004996 | Gadelaidic acid | 30.7 | 0.2 |
| Glycyrrhiza inflata | MOL005000 | Gancaonin G | 60.44 | 0.39 |
| Glycyrrhiza inflata | MOL005001 | Gancaonin H | 50.1 | 0.78 |
| Glycyrrhiza inflata | MOL005003 | Licoagrocarpin | 58.81 | 0.58 |
| Glycyrrhiza inflata | MOL005007 | Glyasperins M | 72.67 | 0.59 |
| Glycyrrhiza inflata | MOL005008 | Glycyrrhiza flavonol A | 41.28 | 0.6 |
| Glycyrrhiza inflata | MOL005012 | Licoagroisoflavone | 57.28 | 0.49 |
| Glycyrrhiza inflata | MOL005013 | 18α-hydroxyglycyrrhetic acid | 41.16 | 0.71 |
| Glycyrrhiza inflata | MOL005016 | Odoratin | 49.95 | 0.3 |
| Glycyrrhiza inflata | MOL005017 | Phaseol | 78.77 | 0.58 |
| Glycyrrhiza inflata | MOL005018 | Xambioona | 54.85 | 0.87 |
| Glycyrrhiza inflata | MOL005020 | Dehydroglyasperins C | 53.82 | 0.37 |
SJZ, Si Jun Zi decoction; OB, oral bioavailability DL, drug-likeness.
Table 2
| Genes | |
|---|---|
| Herb targets & cSCC targets | AR, PPARG, RELA, EGFR, VEGFA, CCND1, BCL2, FOS, CASP9, PLAU, RB1, IL6, CASP3, TP63, NFKBIA, CASP8, RAF1, PRKCA, HIF1A, ERBB2, CAV1, MYC, CYP1A1, ICAM1, SELE, VCAM1, BIRC5, NOS3, HSPB1, CYP1B1, CCNB1, GSTP1, NFE2L2, NQO1, PARP1, CHEK2, CRP, RUNX2, RASSF1, CTSD, IGFBP3, IGF2, IRF1, ERBB3, RASA1, GSTM1, PGR, ESR2, CHEK1, ESR1, GSK3B, IKBKB, MAPK8, ABCC1 |
cSCC, cutaneous squamous cell carcinoma.
Network pharmacology of SJZ decoction
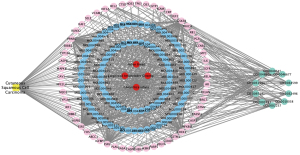
The 54 targets of drug active compounds in SJZ which are related to cSCC are mainly enriched in “response to steroid hormone”, “response to metal ion”, “regulation of apoptotic signaling pathway”, “response to lipopolysaccharide”, “response to molecule of bacterial origin”, “extrinsic apoptotic signaling pathway”, “cellular response to oxidative stress”, “response to antibiotic”, “reproductive structure development”, and “regulation of DNA-binding transcription factor activity” by GO analysis for biological process. In this study, we reintegrated “TCMs-compounds”, “compounds-targets”, “TCMs-targets”, “targets-pathways”, and “disease-targets” by using Cytoscape software and obtained a “TCMs-compounds-targets-pathways-disease” network (Figure 4).
Screening and analysis of cluster and hub genes
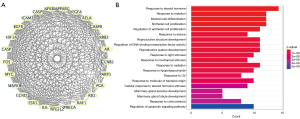
We selected the module with the highest score from 54 common targets through the MCODE plug-in in Cytoscape. This cluster contains the 25 most correlated targets (Figure 5A). The 25 targets are mainly enriched in “response to steroid hormone”, “response to metal ion”, “myeloid cell differentiation”, “epithelial cell proliferation”, and “regulation of epithelial cell proliferation” by GO analysis for biological process (Figure 5B). The CytoHubba plug-in was used to sort degree value as a standard from high to low, and ten genes with the highest degree value were selected (Figure 6). Through GO analysis of hub genes, we found that “mammary gland alveolus development”, “mammary gland lobule development”, “positive regulation of epithelial cell proliferation”, “epithelial cell proliferation”, and “response to light stimulus” were the most closely related biological processes, in which “positive regulation of epithelial cell proliferation” and “epithelial cell proliferation” and Cluster 1 had the same closest relationship. These two processes included the MYC, ESR1, EGFR, HIF1A, CCND1, VEGFA, and ERBB2 genes. The pathways which had the top five highest relevant scores the candidate genes in Cluster 1 were mainly enriched in: “Kaposi sarcoma-associated herpesvirus infection”, “hepatitis B”, “human cytomegalovirus infection”, “chemical carcinogenesis-receptor activation”, and “lipid and atherosclerosis”. The common target-related main signaling pathways might have included: the JNK/NF-κB signaling pathway, the NF-κB/VEFG signaling pathway, the NF-κB/ICAM-1 signaling pathway, the JNK/AP-1 signaling pathway, the AP-1/ICAM-1 signaling pathway, the AP-1/VEFG signaling pathway, the HIF1A/VEFG signaling pathway, and the Caspase-9/Caspase-3 signaling pathway.

UPLC fingerprints
Through comparison with the “Similarity Evaluation System for Chromatographic Fingerprint of TCMs (Version 2012)”, the fingerprints of chromatograms in SJZ decoction showed 13 common peaks, which were identified as Atractylon, Liquiritin, 25-hydroxyporicoic acid H, Isoliquiritin apioside, ginsenoside Rg1, Ginsenoside Rg3, Ginsenoside Re, Kanzonol H, Ginsenoside Ro, Poricoic acid, Licoricesaponin G2, Glycyrrhizic acid, and Liquiritigenin, respectively (Figure 7). These ingredients may provide important laboratory evidence for antitumor therapy of SJZ.

Discussion
As a kind of non-melanoma skin tumor, cSCC has a high incidence and metastasis rate. In the treatment of this disease, surgery is suitable for the early, non-metastatic type, but is not the best choice for metastatic or recurrent cSCC (15). Formulae of TCM have been applied and studied by medical and scientific researchers in many countries worldwide for many years, which have been reported to have the characteristics of high absorption, complex ingredients, and multi-target (16). The different combinations and proportions of various CHMs in the prescription make the drugs have different mechanisms and effects, which has also become a research hotspot in clinical treatment.
The SJZ decoction, as a classical prescription, has been used by many patients to improve the weakness caused by malignant tumors, and is an auxiliary treatment method. In recent years, there are a number of reported results show that SJZ decoction or SJZ based Chinese traditional medicine in the bladder cancer mice, lung cancer mice that accept chemotherapy have enhanced the effect of chemotherapy drugs, reduce the effect of chemotherapy drugs toxic side effects through inhibiting tumor growth and can prolong the survival of mice (17,18). In addition to gastric cancer (19-22), it has also been studied at an animal level or cellular level in the clinical treatment of HOSCC and multiple types of lung cancer (23), but we did not find any previous studies related to SJZ in cSCC.
In this study, the constituents of Ginseng, Glycyrrhiza inflata, Largehead Atractylodes, and Indian Bread in SJZ decoction were screened by network pharmacology, and the targets of these candidate constituents were further analyzed and categorized. At the same time, comprehensive comparison and intersection were made between these potential targets above and potential targets related to cSCC, and candidate targets were screened out for functional enrichment analysis. Meanwhile, a network diagram of “drug-component-target-pathway-disease” was made for future reference and use.
The study found that the potential targets of the more prominent active ingredients of SJZ decoction were mainly concentrated in the biological processes: response to steroid hormone and may be most closely related to the pathways: Kaposi sarcoma-associated herpesvirus infection. At the same time, these targets, pathways, and biological processes are related to the occurrence and development of cSCC.
The results of UPLC analysis showed that the top ten active ingredients extracted from SJZ decoction were Atractylon, Liquiritin, 25-hydroxyporicoic acid H, Isoliquiritin apioside, ginsenoside Rg1, Ginsenoside Rg3, Ginsenoside Re, Kanzonol H, Ginsenoside Ro, Poricoic acid, Licoricesaponin G2, Glycyrrhizic acid, and Liquiritigenin. The ingredients come from the four different Chinese herbs.
In short, the ten main active components of SJZ decoction analyzed by UPLC may act on cSCC-related targets through specific pathways, and then play a role in the treatment or adjuvant treatment of cSCC.
Conclusions
This study conducted correlation analysis on the active components of SJZ decoction and the disease target of cSCC through network pharmacology, and constructed a network diagram with software combining TCM-component-target-disease-pathway. The analysis of the effective components of SJZ decoction by UPLC is helpful to provide a reference direction and theoretical basis for omgoing research on the treatment of cSCC by SJZ decoction.
Limitation
Whether SJZ decoction has the effect of preventing and treating cancer and its mechanism of action still need to be further explored.
Acknowledgments
Funding: This work was supported by Project of Administration of Traditional Chinese Medicine of Guangdong Province, China (No. 20221014) and supported by The Science Foundation of Guangdong Second Provincial General Hospital (YQ2019-002).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1716/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fania L, Didona D, Di Pietro FR, et al. Cutaneous Squamous Cell Carcinoma: From Pathophysiology to Novel Therapeutic Approaches. Biomedicines 2021;9:171. [Crossref] [PubMed]
- Keeping S, Xu Y, Chen CI, et al. Comparative efficacy of cemiplimab versus other systemic treatments for advanced cutaneous squamous cell carcinoma. Future Oncol 2021;17:611-27. [Crossref] [PubMed]
- Hughes BGM, Munoz-Couselo E, Mortier L, et al. Pembrolizumab for locally advanced and recurrent/metastatic cutaneous squamous cell carcinoma (KEYNOTE-629 study): an open-label, nonrandomized, multicenter, phase II trial. Ann Oncol 2021;32:1276-85. [Crossref] [PubMed]
- Keohane SG, Botting J, Budny PG, et al. British Association of Dermatologists guidelines for the management of people with cutaneous squamous cell carcinoma 2020. Br J Dermatol 2021;184:401-14. [Crossref] [PubMed]
- Unlu A, Nayir E, Kirca O, et al. Ginseng and cancer. J BUON 2016;21:1383-7. [PubMed]
- Sabouri-Rad S, Sabouri-Rad S, Sahebkar A, et al. Ginseng in Dermatology: A Review. Curr Pharm Des 2017;23:1649-66. [Crossref] [PubMed]
- Huang HC, Tsai LL, Tsai JP, et al. Licochalcone A inhibits the migration and invasion of human lung cancer cells via inactivation of the Akt signaling pathway with downregulation of MMP-1/-3 expression. Tumour Biol 2014;35:12139-49. [Crossref] [PubMed]
- Kang TH, Seo JH, Oh H, et al. Licochalcone A Suppresses Specificity Protein 1 as a Novel Target in Human Breast Cancer Cells. J Cell Biochem 2017;118:4652-63. [Crossref] [PubMed]
- Kim KH, Yoon G, Cho JJ, et al. Licochalcone A induces apoptosis in malignant pleural mesothelioma through downregulation of Sp1 and subsequent activation of mitochondria-related apoptotic pathway. Int J Oncol 2015;46:1385-92. [Crossref] [PubMed]
- Wu CP, Lusvarghi S, Hsiao SH, et al. Licochalcone A Selectively Resensitizes ABCG2-Overexpressing Multidrug-Resistant Cancer Cells to Chemotherapeutic Drugs. J Nat Prod 2020;83:1461-72. [Crossref] [PubMed]
- Yuan LW, Jiang XM, Xu YL, et al. Licochalcone A inhibits interferon-gamma-induced programmed death-ligand 1 in lung cancer cells. Phytomedicine 2021;80:153394. [Crossref] [PubMed]
- Oh HN, Lee MH, Kim E, et al. Licochalcone B inhibits growth and induces apoptosis of human non-small-cell lung cancer cells by dual targeting of EGFR and MET. Phytomedicine 2019;63:153014. [Crossref] [PubMed]
- Oh HN, Lee MH, Kim E, et al. Licochalcone D Induces ROS-Dependent Apoptosis in Gefitinib-Sensitive or Resistant Lung Cancer Cells by Targeting EGFR and MET. Biomolecules 2020;10:297. [Crossref] [PubMed]
- Oh HN, Seo JH, Lee MH, et al. Licochalcone C induced apoptosis in human oral squamous cell carcinoma cells by regulation of the JAK2/STAT3 signaling pathway. J Cell Biochem 2018;119:10118-30. [Crossref] [PubMed]
- Kwak AW, Cho SS, Yoon G, et al. Licochalcone H Synthesized by Modifying Structure of Licochalcone C Extracted from Glycyrrhiza inflata Induces Apoptosis of Esophageal Squamous Cell Carcinoma Cells. Cell Biochem Biophys 2020;78:65-76. [Crossref] [PubMed]
- Nho SH, Yoon G, Seo JH, et al. Licochalcone H induces the apoptosis of human oral squamous cell carcinoma cells via regulation of matrin 3. Oncol Rep 2019;41:333-40. [PubMed]
- Liu Y, Jia Z, Dong L, et al. A randomized pilot study of atractylenolide I on gastric cancer cachexia patients. Evid Based Complement Alternat Med 2008;5:337-44. [Crossref] [PubMed]
- Ye Yan, Chou GX, Wang Hui, et al. Effects of sesquiterpenes isolated from largehead atractylodes rhizome on growth, migration, and differentiation of B16 melanoma cells. Integr Cancer Ther 2011;10:92-100. [Crossref] [PubMed]
- Knackstedt TJ, Knackstedt RW, Djohan M, et al. New Developments in the Management of Cutaneous Squamous Cell Carcinoma. Plast Reconstr Surg 2021;147:492-504. [Crossref] [PubMed]
- Wang L, Xu L, Wang Y. Huaier Inhibits Proliferation, Migration, and Invasion of Cutaneous Squamous Cell Carcinoma Cells by Inhibiting the Methylation Levels of CDKN2A and TP53. Integr Cancer Ther 2021;20:15347354211031646. [Crossref] [PubMed]
- Li YJ, Liao LL, Liu P, et al. Sijunzi Decoction Inhibits Stemness by Suppressing beta-Catenin Transcriptional Activity in Gastric Cancer Cells. Chin J Integr Med 2021; [Crossref] [PubMed]
- Ding P, Guo Y, Wang C, et al. A Network Pharmacology Approach for Uncovering the Antitumor Effects and Potential Mechanisms of the Sijunzi Decoction for the Treatment of Gastric Cancer. Evid Based Complement Alternat Med 2022;2022:9364313. [Crossref] [PubMed]
- Shao N, Xiao Y, Zhang J, et al. Modified Sijunzi Decoction Inhibits Epithelial-Mesenchymal Transition of Non-Small Cell Lung Cancer by Attenuating AKT/GSK3beta Pathway in vitro and in vivo. Front Pharmacol 2022;12:821567. [Crossref] [PubMed]
(English Language Editor: J. Jones)


