
Development and validation of a combined ferroptosis and immune prognostic signature for lung adenocarcinoma
Introduction
Lung cancer is one of the most common malignancies worldwide and is responsible for the leading cause of cancer-related deaths, accounting for 18% of all cancer deaths in 2020 (1). Lung adenocarcinoma (LUAD) accounts for 40% of all subtypes of lung cancer (2). Research and treatment for LUAD have advanced significantly, but the outcomes for LUAD patients remain poor (3). Generally, although in tumors with similar clinical and pathological characteristics, there is great heterogeneity in the clinical outcomes and therapy responses of patients. In recent years, there has been increasing evidence that variations in patient genomes contribute significantly to interpatient differences in drug response (4,5). Therefore, more and more studies are attempting to identify subgroups of tumor patients based on their molecular profiles (6,7). In the past decade, with the rapid development of high-throughput sequencing, gene set has shown great potential in personalized therapy. Specific gene set has led to a better understanding of the biological phenotypes of cancer than single biomarkers (8).
Ferroptosis is a new form of cell death and is mainly characterized by iron-dependent lipid peroxide accumulation and reactive oxygen species production (9). There is growing evidence that ferroptosis is associated with many neurodegenerative diseases such as Alzheimer’s disease (10), Parkinson’s disease (11), heart disease (12), blood diseases (13,14), and immune system disorders (15). Also, many types of cancer cells are highly sensitive to ferroptosis, including renal cancer (16), breast cancer (17), human non-small cell lung cancer, and colorectal cancer cells (18). New evidence suggested that ferroptosis was inhibited in a variety of tumors, and thus the regulation of ferroptosis could be a new approach to cancer treatment (19). Ferroptosis elicits therapeutic responses that inhibit tumor growth when used experimental reagents, approved drugs, ionizing radiation, and cytokines (20). Studies have shown that dysregulation of ferroptosis may contribute to lung cancer development (21,22). Numerous studies have explored the relationship between certain ferroptosis-related genes (FRGs) and lung cancer. Lai et al. (23) found that glutathione peroxidase 4 (GPX4) overexpressed in non-small cell lung cancer (NSCLC) tissues and cell lines, and the upregulation of GPX4 reduced multiple mitochondrial abnormalities which is one of the features of ferroptosis and promoted proliferation of lung cancer cells. Moreover, cystine/glutamate antiporter (SLC7A11), regulating metabolic demand during the development of NSCLC, was upregulated in NSCLC tissues and was associated with a poorer 5-year survival rate (24). Huang et al. (25) found that in LUAD cells (A549 cells), p53 activation by erastin exposure induced ferroptosis and apoptosis and arrested the cell cycle at the G1 phase, thereby inhibited cell proliferation. Therefore, for LUAD, targeting ferroptosis may be beneficial to explore more effective anticancer therapies.
Abnormal ferroptosis is closely associated with a dysregulated immune response. An important study found that activation of CD8+ T cells by immunotherapy resulted in the release of interferon (IFN)-γ to downregulate the expression of SLC3A2 and SLC7A11, which in turn enhanced ferroptosis-specific lipid peroxidation (26). Therefore, targeting this pathway is beneficial in increasing antitumor efficacy. In addition, the interactions among ferroptosis, inflammation, and the immune system in the tumor microenvironment (TME) influence tumor progression. The synergy of ferroptosis and immunity not only inhibits primary tumor but also stimulates immune responses by combining with immune checkpoint inhibitors (ICIs) (27). In conclusion, exploring the integrated regulatory network of ferroptosis and immunity in LUAD might provide clues to new anti-cancer strategies.
In this study, the combined signature of ferroptosis- and immune-related genes (CSFI) was constructed using the Cancer Genome Atlas (TCGA) database. Patients with LUAD from the Gene Expression Omnibus (GEO) database were applied for the validation set. The relationship between CSFI and survival status was explored and a nomogram was developed to predict the survival rates of LUAD patients. Finally, the immune and ferroptosis status of different CSFI subgroups were evaluated to guide anti-tumor personalized therapy. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-992/rc).
Methods
Data acquisition
RNA-seq data of 535 LUAD patients and 59 normal patients were obtained from TCGA (https://portal.gdc.cancer.gov/). The corresponding clinical data were acquired from UCSC Xena (http://xena.ucsc.edu/). Patients with incomplete survival information were excluded from further analyses. Meanwhile, mRNA expression data and clinical information of 3 LUAD datasets (GSE31210, GSE50081, and GSE42127) were downloaded from the GEO repository (https://www.ncbi.nlm.nih.gov/geo/). The 3 datasets are combined as the validation dataset and the “sva” package was applied to remove the batch effect. Additionally, 259 FRGs were obtained from the FerrDb website (http://www.datjar.com:40013/bt2104/) and 2,483 immune-related genes (IRGs) were acquired from the ImmPort website (https://www.immport.org/) (available online: https://cdn.amegroups.cn/static/public/tcr-22-992-1.pdf). These FRGs were classified into three categories according to the regulatory effects of FRGs on ferroptosis, namely 163 drivers of ferroptosis (DOFs), 84 suppressors of ferroptosis (SOFs), and the remaining 12 either promote or suppress ferroptosis (Table S1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Generation of CSFI
To assess the combined effect of ferroptosis and immunity on LUAD, CSFI which was composed of key FRGs and IRGs was developed. The screening process for these key genes was as follows. Firstly, differentially expressed genes (DEGs) between tumor samples and normal samples were identified by the “limma” package with the absolute values of log-fold change (|logFC|) ≥1 and false discovery rate (FDR) <0.05. “Venn” package in R was applied to identify differentially expressed FRGs and IRGs. Secondly, these differentially expressed FRGs and IRGs were used for univariate Cox regression to identify prognostic genes. FRGs with P<0.05 and IRGs with P<0.01 were regarded as prognostic genes. The protein-protein interaction (PPI) network of these prognostic genes was constructed through the STRING database (https://www.string-db.org/). Cytoscape_v3.8.2 was used for visualization. Thirdly, these prognostic genes were used to construct the Least Absolute Shrinkage and Selection Operator (LASSO)-Cox regression model. Finally, several key genes and their coefficients were retained through the R package “glmnet”. Meanwhile, their expression levels and coefficients were used for calculating the values of CSFI, namely , X: coefficients, Y: values of gene expression).
Clinical benefit assessment
Then, LUAD patients from TCGA were divided into CSFI-high and CSFI-low groups based on the median CSFI values. Principal component analysis (PCA) was performed to assess the clustering efficacy of CSFI. For appraising the sensitivity and specificity of prediction, the time-dependent receiver operating characteristic (ROC) curves were executed. According to the formula derived from the TCGA cohort, the CSFI values of patients in the validation cohort were calculated to further validate the results from the TCGA cohort. To validate the independent prognostic value of CSFI, univariate and multivariate Cox regression analyses were performed in the TCGA cohort. Last but not least, the nomogram model was constructed based on multivariate Cox regression results. Moreover, the concordance index (C-index) was calculated and calibration curves were plotted to evaluate the performance of the nomogram with the “rms” package of R.
Ferroptosis profile and functional enrichment analyses
SOFs and DOFs validated by human and rat experiments were extracted for the construction of the PPI network respectively. And Cytoscape software was used to filter the top 10 SOFs and DOFs based on degree value. The expressions of these genes were compared in different CSFI groups.
Gene Ontology (GO) and Kyoto Gene and Genome Encyclopedia (KEGG) analyses were carried out to excavate the function of DEGs between CSFI-high and CSFI-low groups by applying the “clusterProfiler” R package. Meanwhile, Gene Set Enrichment Analysis (GSEA) was performed with the reference gene set (c2.cp.v7.1.symbols.gmt) to explore pathways significantly enriched in two CSFI groups through GSEA (version 4.2.2). Pathways with nominal P value <0.05, FDR q-value <0.25, and normalized enrichment score (NES) >1 were regarded to be significantly enriched.
Mutation analysis and evaluation of immune cell infiltration
The somatic mutation data of LUAD patients were downloaded from the TCGA database. The “maftools” package in R was used for analysis and visualization (pipeline: mutect2). The expression levels of 6 immune checkpoint genes (ICPGs) (CD274, HAVCR2, IDO1, LAG3, PDCD1, PDCD1LG2) between CSFI-high and CSFI-low groups were compared. Additionally, single-sample gene set enrichment analysis (ssGSEA) was employed to calculate scores of immune cells and immune-related pathways in different CSFI groups by “gsva” packages in R. All signatures used in our study were listed in website: https://cdn.amegroups.cn/static/public/tcr-22-992-2.pdf.
Statistical analysis
Kaplan-Meier (KM) method with the log-rank test was employed for comparing the survival status of different CSFI groups. Time-dependent ROC curve analyses were performed using “pROC” using packages in R. The “survminer and “survival” were applied for univariate and multivariate Cox regression analysis. Mann-Whitney U test was used to compare scores of immune cells and immune-related pathways, and ICPGs expression between CSFI-high and CSFI-low groups. All statistical analyses were carried out with R software (version 4.1.0). A value of P<0.05 was considered to be statistically significant. The flow diagram is shown in Figure 1.
Results
Construction of CSFI in LUAD
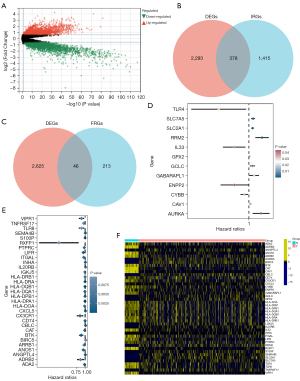
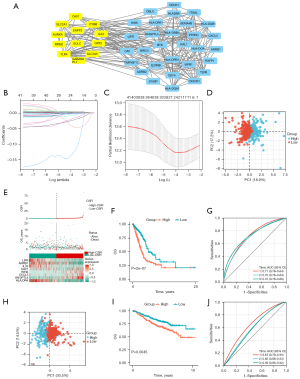
Based on analysis of gene expression differences, 2,671 genes were regarded as DEGs between tumor and normal samples. Among them, 1,628 genes were downregulated while 1,043 were upregulated (Figure 2A). As shown in Figure 2B,2C, 378 differentially expressed IRGs and 46 differentially expressed FRGs were identified for performing univariate Cox regression analysis. Then, 12 FRGs and 31 IRGs were proved to be significantly related to the prognoses of LUAD patients (all P<0.05) (Figure 2D,2E). The expression levels of these prognostic genes are presented in Figure 2F. In addition, as shown in Figure 3A, there was a strong relationship between prognosis-related FRGs and IRGs. Based on these prognostic genes, the LASSO regression and multivariate Cox regression were performed to construct an eight-gene signature (CSFI) model to predict the survival outcomes of LUAD patients (Figure 3B,3C). The value of CSFI were computed via formula: CSFI = (0.0052 × ANGPTL4 exp.) + (–0.0260 × ARRB1 exp.) + (0.0030 × CAV1 exp.) + (0.0063 × CXCL5 exp.) + (-0.0003 × HLA-DRA exp.) + (-0.0135 × IL33 exp.) + (0.0052 × INHA exp.) + (-0.0469 × LIFR exp.). Among them, CAV1 and IL33 belonged to FRGs, and ANGPTL4, ARRB1, CXCL5, HLA-DRA, IL33, INHA, and LIFR were the parts of IRGs. According to the median CSFI value, LUAD patients in the TCGA cohort were divided into CSFI-high (N=261) and CSFI-low (N=261) groups. PCA showed patients of the two groups were distinctively clustered (Figure 3D). It indicated that CSFI could well distinguish the two CSFI groups. Figure 3E presents the distribution of CSFI, overall survival (OS), and gene expression levels of eight genes in two groups. KM curves revealed that patients in the CSFI-high group have poorer prognoses instead of those in the CSFI-low group (log-rank P<0.001) (Figure 3F). To appraise the sensitivity and specificity of prediction of the CSFI, 1–3-year time-dependent ROC curves were plotted and area under curve (AUC) values were calculated. The 1–3-year survival AUC was 0.71, 0.70, and 0.72 respectively (Figure 3G). For the validation cohort, CSFI values of LUAD patients were calculated based on a formula derived from the TCGA cohort. Similarly, two CSFI groups manifested obvious separation through PCA analysis (Figure 3H). KM analysis indicated that the CSFI-high group had a lower survival rate than the CSFI-low group (P=0.0045) (Figure 3I). As shown in Figure 3J, the AUCs were 0.67 for 1-year, 0.60 for 2-year, and 0.59 for 3-year.
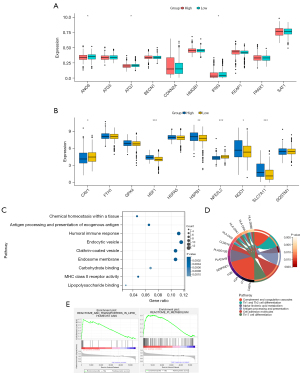
Ferroptosis profile and functional enrichment analyses in different CSFI groups
To explore the ferroptosis status of patients with different CSFI values, the transcriptional changes of 20 SOFs and DOFs were studied in the CSFI subgroups of the TCGA cohort. It was found that several DOFs, including ANO6, ATG7, and IFNG, were downregulated in the CSFI-high group (P<0.05) (Figure 4A). Moreover, some SOFs, including HSF1, HSPB1, NQO1, and SLC7A11, were upregulated in the CSFI-high group (P<0.05), while CAV1 and NFE2L2 were downregulated in CSFI-high group (P<0.05) (Figure 4B). From this, it can be speculated that there was a ferroptosis-inhibited status in the CSFI-high group. According to GO (Figure 4C) and KEGG analyses (Figure 4D), DEGs between CSFI-high and CSFI-low groups were correlated with chemical homeostasis, alpha-linolenic acid metabolism, lipopolysaccharide binding, transport of vesicle, and other immune responses such as antigen processing and presentation. Additionally, the results of GSEA showed that genes in the CSFI-low group were significantly enriched in ABC transporters in lipid homeostasis and phosphatidylinositol metabolism (Figure 4E). Therefore, it is more favorable to consider the presence of ferroptosis inhibition in the CSFI-high group.
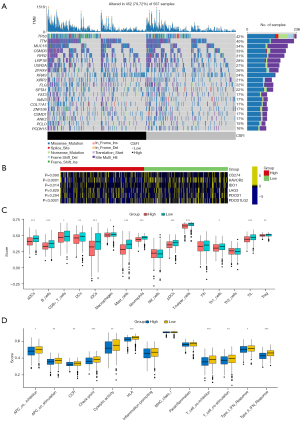
Gene mutation and immune profiles in different CSFI groups
Figure 5A showed the gene mutation profiles of different CSFI groups in the TCGA cohort. It was found that the top 5 genes with the highest mutation frequencies in LUAD patients were TP53 (42%), TTN (40%), MUNC16 (34%), CSMD3 (33%), and RYR2 (31%). And the missense mutation was the main type of mutation. Furthermore, it was obvious that the mutation frequencies of TP53 in the CSFI-high group were higher than CSFI-low group. To precisely guide immunotherapy, the expressions of 6 ICPGs were compared between CSFI-high and -low groups (Figure 5B). It was shown that patients in the CSFI-high group had lower expressions of HAVCR2 (P<0.0001), IDO1 (P=0.014), and PDCD1LG2 (P<0.0001) rather than those in the CSFI-low group. In addition, scores of 16 types of immune cells and immune-related pathways were compared between CSFI-high and -low groups in the TCGA cohort through performing ssGSEA. Firstly, because scores of dendritic cells (DCs), activated dendritic cells (aDCs), induced dendritic cells (iDCs), plasmacytoid dendritic cells (pDCs), B cells, CD8+ T cells, Macrophages, Mast cells, Neutrophils, T helper (Th) cells, Th1 cells, tumour-infiltrating lymphocytes (TILs), and regulatory T cells (Tregs) were lower in the CSFI-high group rather than CSFI-low group (all P<0.05) (Figure 5C). It was reasonable to assume that the CSFI-high group was equipped with lower levels of immune cell infiltration. Moreover, it was proved that the CSFI-high group had lower activity of immune-related pathways. The scores of the other 10 types of immune-related pathways were lower in CSFI-high patients than in CSFI-low patients (all P<0.05) (Figure 5D), excluding inflammation promoting, major histocompatibility complex (MHC) class I, and parainflammation.
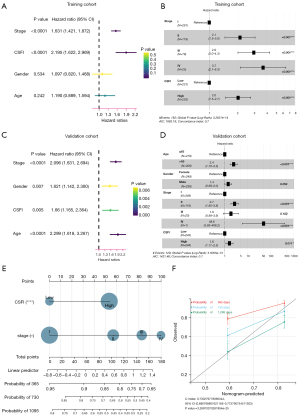
Identification of independent prognostic value of the CSFI
Firstly, for the TCGA cohort, stage and CSFI were proved to be risk factors for the prognosis of LUAD (P<0.001) by univariate Cox analysis (Figure 6A). Subsequently, after multivariate Cox analysis, stage and CSFI [hazard ratio (HR) =2.0, 95% confidence interval (CI): 1.5–2.7, P<0.001] were still prognostic predictors (Figure 6B). Then, in the validation cohort, age, stage, and CSFI (HR =1.6, 95% CI: 1.11–2.3, P=0.011) were independent risk factors by employing univariate (Figure 6C) and multivariate (Figure 6D) Cox regression analyses. Last but not least, a nomogram was constructed for predicting the survival rates (Figure 6E). The C-index of model was 0.73 (95% CI: 0.69–0.77, P<0.001). The calibration plots suggested that the model appeared good predictive validity in 2- and 3-year survival rates (Figure 6F).
Discussion
In the past decade, patients with LUAD have seen some improvement in the quality of their survival, all thanks to targeted therapies and immunotherapy. However, only a small percentage of patients could get effective treatment from targeted therapies and immunotherapy. Therefore, it is necessary and urgent to explore new anti-cancer therapies to benefit more patients. In recent years, the development of anti-cancer therapies targeting specific molecular profile alterations or phenotypic characteristics has become an emerging trend. This treatment is based on molecular markers and provides personalized treatment for every patient (5,8,28). For example, in one study, seven pyroptosis-related genes were used to classify patients with ovarian cancer into high- and low-risk groups. And ultimately it was found that patients in the high-risk group had a state of immunosuppression (29). Moreover, the hypoxia-immune signature which was composed of ten hypoxia-related or immune-related genes divided patients with triple-negative breast cancer into two subgroups. It was proved that immunotherapy may be more effective in patients in the hypoxia low/immune high group (7). A growing number of studies are now confirming the interaction between ferroptosis and immunity in tumors (20,26,30). On the one hand, CD8+ T cells released IFN-γ, which can activate the JAK1-STAT1 signaling pathway. And at last induced ferroptosis in tumor cells by regulating the expression of SLC7A11 and SLC3A2. In addition, transforming growth factor β1 (TGF-β1) released by macrophages regulated FRG expressions by activating small mothers against decapentaplegic (SMAD) protein-mediated signaling pathways. On the other hand, the release of damage-associated molecular patterns (DAMPs) from ferroptosis cancer cells affected the function of innate immune cells in TME (20).
Considering the close relationship between the immune system and ferroptosis, CSFI was constructed based on IRGs and FRGs in this study. According to the training and validation cohort, the OSs of patients in the CSFI-high group were significantly shorter than those in the CSFI-low group. In addition, combining the expressions of some SOFs and DOFs, it can be speculated that CSFI-high patients had a state of ferroptosis inhibition. In this study, the expressions of several DOFs (ANO6, ATG7, and IFNG), which can contribute to ferroptosis in cancer cells were significantly downregulated in the CSFI-high group (31-33). In addition, some SOFs, including HSF1, HSPB1, NQO1, and SLC7A11, were upregulated in the CSFI-high group. SLC7A11, a well-defined inhibitor of ferroptosis, is a key pathway for many tumor suppressor genes to inhibit tumor formation. For instance, BAP1 and p53 inhibited SLC7A11 to induce ferroptosis, while KEAP1 prevented NRF2 from activating SLC7A11 (34-36). An important finding showed that YTHDC2, which can serve as a ferroptosis inducer targeted system XC- for the treatment of LUAD (37). Then, the low expressions of NRF2 and NQO1 predicted better survival outcomes in NSCLC patients (38). Furthermore, Sun et al. (39) found that the inhibition of the HSF1-HSPB1 pathway increased the anti-cancer activity of erastin. And cells were more susceptible to ferroptosis if the HSBP1-HSF1 pathway was inhibited when cancer cells contained high levels of iron and HSPB1. In NSCLC, researchers found that the knockdown of HSF1 during co-treatment with erastin and celastrol inhibited HSP to increase cell death. The combined treatment of HSF1 inhibition and chemotherapy might be a new target for NSCLC treatment (40). In this study, the upregulation of HSF1, HSPB1, NQO1, and SLC7A11 means that patients from the CSFI-high group might be equipped with strong ferroptosis resistance and the function of ferroptosis inducers might be diminished. This is also the reason for the poor prognosis. Therefore, the therapy by combining ferroptosis inducers and targeting SLC7A11, HSF1, and HSPB1 might result in better outcomes in the CSFI-high group. Moreover, it has been found that there was an interaction between lipid metabolism and ferroptosis in the development and progression of cancer (41). The lipid metabolism is closely related to ferroptosis (42). Based on GSEA analysis, genes in the CSFI-low group were significantly enriched in ABC transporters in lipid homeostasis and phosphatidylinositol metabolism. GO and KEGG analyses also manifested DEGs between different CSFI groups enriched in some pathways, such as lipopolysaccharide binding, chemical homeostasis, and alpha-linolenic acid metabolism. It indicated that the CSFI-low group was likely to be sensitive to ferroptosis inducers.
LUAD patients with high CSFI values appeared immune suppressive state. ssGSEA analysis results showed that scores of several dendritic cells, mast cells, B cells, CD8+ T cells, macrophages, neutrophils, Th1 cells, TILs, and Tregs were lower in CSFI-high group patients. Scientific evidence suggested that mast cells, B cells, CD8 + T cells, and TILs were related to the good prognosis of lung cancer (43-45). There were also studies showing that DCs and Th1 cells induce antitumor immunity in lung cancer patients (46,47). Additionally, although macrophages promote tumor growth and are associated with poor prognosis in most advanced tumors, there is growing evidence that anti-tumor immune responses are greatly enhanced by the appropriate activation of macrophages (48). And neutrophils exert pro- or anti-tumor effects, depending on different subpopulations (49). Moreover, in NSCLC, Tregs were positively associated with tumor metastasis and poor survival (50). However, research published in “Nature Immunology” found that Type 1 Treg enhanced immune response (51). In this study, the level of Treg was higher in CSFI-low group patients with better prognoses. Consequently, it is necessary to identify the Treg isoforms that play a role in LUAD. In summary, it could be concluded that the poor prognoses of patients in the CSFI-high group might be due to impaired immune function and poor anti-tumor immune response. A previous study found that NSCLC patients generated a significant and durable response to ICIs’ treatment (52). By analyzing the expression of ICPGs in both groups, it could be hypothesized that immunotherapies against HAVCR2, IDO1, and PDL2 were effective on patients of the CSFI-low group.
TP53 encodes p53 protein and thus exerts tumor-suppressive effects (53). TP53 mutation occurs most frequently and is often associated with a worse prognosis in a variety of human cancers, including LUAD (54). In this study, the frequencies of TP53 mutation were higher in patients with higher CSFI values. In particular, abnormal p53 contributes to immune escape, resulting in an immunosuppressive microenvironment (55). It has been shown that TP53 mutation could lead to reduced immunity in LUAD patients (56). As a result, higher frequencies of TP53 mutation in patients in the CSFI-high group might have contributed to the immunosuppressed state of these patients.
More and more studies are exploring the association between ferroptosis and immunity, but there was no study that combined FRGs with IRGs to construct a model to predict OSs in LUAD patients. In this study, the prognoses of LUAD patients were more accurately predicted by the nomogram constructed based on tumor stage and CSFI. And based on the LASSO-cox model, more targeted treatment directions were provided for patients in both CSFI subgroups. However, this study is still based on retrospective data, and the predictive value of the model requires prospective studies for multiple validations.
Conclusions
In conclusion, the CSFI constructed in this study could predict the prognoses of LUAD patients robustly and provide targeted treatment directions based on the phenotypic characteristics of different subgroups of patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-992/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-992/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-992/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Meza R, Meernik C, Jeon J, et al. Lung cancer incidence trends by gender, race and histology in the United States, 1973-2010. PLoS One 2015;10:e0121323. [Crossref] [PubMed]
- Hutchinson BD, Shroff GS, Truong MT, et al. Spectrum of Lung Adenocarcinoma. Semin Ultrasound CT MR 2019;40:255-64. [Crossref] [PubMed]
- Lee JW, Aminkeng F, Bhavsar AP, et al. The emerging era of pharmacogenomics: current successes, future potential, and challenges. Clin Genet 2014;86:21-8. [Crossref] [PubMed]
- Abubakar MB, Gan SH. Molecular Targets in Advanced Therapeutics of Cancers: The Role of Pharmacogenetics. Oncology 2016;91:3-12. [Crossref] [PubMed]
- Yin X, Wang Z, Wang J, et al. Development of a novel gene signature to predict prognosis and response to PD-1 blockade in clear cell renal cell carcinoma. Oncoimmunology 2021;10:1933332. [Crossref] [PubMed]
- Zheng S, Zou Y, Liang JY, et al. Identification and validation of a combined hypoxia and immune index for triple-negative breast cancer. Mol Oncol 2020;14:2814-33. [Crossref] [PubMed]
- Tsimberidou AM, Fountzilas E, Nikanjam M, et al. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat Rev 2020;86:102019. [Crossref] [PubMed]
- Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. [Crossref] [PubMed]
- Cong L, Dong X, Wang Y, et al. On the role of synthesized hydroxylated chalcones as dual functional amyloid-β aggregation and ferroptosis inhibitors for potential treatment of Alzheimer's disease. Eur J Med Chem 2019;166:11-21. [Crossref] [PubMed]
- Mahoney-Sánchez L, Bouchaoui H, Ayton S, et al. Ferroptosis and its potential role in the physiopathology of Parkinson's Disease. Prog Neurobiol 2021;196:101890. [Crossref] [PubMed]
- Kobayashi M, Suhara T, Baba Y, et al. Pathological Roles of Iron in Cardiovascular Disease. Curr Drug Targets 2018;19:1068-76. [Crossref] [PubMed]
- Martinet W, Coornaert I, Puylaert P, et al. Macrophage Death as a Pharmacological Target in Atherosclerosis. Front Pharmacol 2019;10:306. [Crossref] [PubMed]
- Probst L, Dächert J, Schenk B, et al. Lipoxygenase inhibitors protect acute lymphoblastic leukemia cells from ferroptotic cell death. Biochem Pharmacol 2017;140:41-52. [Crossref] [PubMed]
- Wang D, Xie N, Gao W, et al. The ferroptosis inducer erastin promotes proliferation and differentiation in human peripheral blood mononuclear cells. Biochem Biophys Res Commun 2018;503:1689-95. [Crossref] [PubMed]
- Yang WS. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. [Crossref] [PubMed]
- Ma S, Henson ES, Chen Y, et al. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis 2016;7:e2307. [Crossref] [PubMed]
- Guo J, Xu B, Han Q, et al. Ferroptosis: A Novel Anti-tumor Action for Cisplatin. Cancer Res Treat 2018;50:445-60. [Crossref] [PubMed]
- Wang H, Cheng Y, Mao C, et al. Emerging mechanisms and targeted therapy of ferroptosis in cancer. Mol Ther 2021;29:2185-208. [Crossref] [PubMed]
- Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 2021;18:280-96. [Crossref] [PubMed]
- Li G, Yang J, Zhao G, et al. Dysregulation of ferroptosis may involve in the development of non-small-cell lung cancer in Xuanwei area. J Cell Mol Med 2021;25:2872-84. [Crossref] [PubMed]
- Zou J, Wang L, Tang H, et al. Ferroptosis in Non-Small Cell Lung Cancer: Progression and Therapeutic Potential on It. Int J Mol Sci 2021;22:13335. [Crossref] [PubMed]
- Lai Y, Zhang Z, Li J, et al. STYK1/NOK correlates with ferroptosis in non-small cell lung carcinoma. Biochem Biophys Res Commun 2019;519:659-66. [Crossref] [PubMed]
- Ji X, Qian J, Rahman SMJ, et al. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene 2018;37:5007-19. [Crossref] [PubMed]
- Huang C, Yang M, Deng J, et al. Upregulation and activation of p53 by erastin induced reactive oxygen species contribute to cytotoxic and cytostatic effects in A549 lung cancer cells. Oncol Rep 2018;40:2363-70. [Crossref] [PubMed]
- Wang W, Green M, Choi JE, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. [Crossref] [PubMed]
- Li Z, Rong L. Cascade reaction-mediated efficient ferroptosis synergizes with immunomodulation for high-performance cancer therapy. Biomater Sci 2020;8:6272-85. [Crossref] [PubMed]
- Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer 2015;137:262-6. [Crossref] [PubMed]
- Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov 2021;7:71. [Crossref] [PubMed]
- Xu H, Ye D, Ren M, et al. Ferroptosis in the tumor microenvironment: perspectives for immunotherapy. Trends Mol Med 2021;27:856-67. [Crossref] [PubMed]
- Lang X, Green MD, Wang W, et al. Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer Discov 2019;9:1673-85. [Crossref] [PubMed]
- Ousingsawat J, Schreiber R, Kunzelmann K. TMEM16F/Anoctamin 6 in Ferroptotic Cell Death. Cancers (Basel) 2019;11:625. [Crossref] [PubMed]
- Zhou B, Liu J, Kang R, et al. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol 2020;66:89-100. [Crossref] [PubMed]
- Stockwell BR, Jiang X, Gu W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol 2020;30:478-90. [Crossref] [PubMed]
- Zhang Y, Shi J, Liu X, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 2018;20:1181-92. [Crossref] [PubMed]
- Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 2013;27:2179-91. [Crossref] [PubMed]
- Ma L, Zhang X, Yu K, et al. Targeting SLC3A2 subunit of system XC- is essential for m6A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic Biol Med 2021;168:25-43. [Crossref] [PubMed]
- Tong YH, Zhang B, Yan YY, et al. Dual-negative expression of Nrf2 and NQO1 predicts superior outcomes in patients with non-small cell lung cancer. Oncotarget 2017;8:45750-8. [Crossref] [PubMed]
- Sun X, Ou Z, Xie M, et al. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 2015;34:5617-25. [Crossref] [PubMed]
- Liu M, Fan Y, Li D, et al. Ferroptosis inducer erastin sensitizes NSCLC cells to celastrol through activation of the ROS-mitochondrial fission-mitophagy axis. Mol Oncol 2021;15:2084-105. [Crossref] [PubMed]
- Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther 2020;5:108. [Crossref] [PubMed]
- Zhang Y, Guo R, Li J, et al. Research progress on the occurrence and therapeutic mechanism of ferroptosis in NSCLC. Naunyn Schmiedebergs Arch Pharmacol 2022;395:1-12. [Crossref] [PubMed]
- Varricchi G, Galdiero MR, Loffredo S, et al. Are Mast Cells MASTers in Cancer? Front Immunol 2017;8:424. [Crossref] [PubMed]
- Xiong L, Wei Y, Zhou X, et al. AGTR1 Inhibits the Progression of Lung Adenocarcinoma. Cancer Manag Res 2021;13:8535-50. [Crossref] [PubMed]
- Brambilla E, Le Teuff G, Marguet S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:1223-30. [Crossref] [PubMed]
- Wang JB, Huang X, Li FR. Impaired dendritic cell functions in lung cancer: a review of recent advances and future perspectives. Cancer Commun (Lond) 2019;39:43. [Crossref] [PubMed]
- Lee H, Choi H. Investigating the Clinico-Molecular and Immunological Evolution of Lung Adenocarcinoma Using Pseudotime Analysis. Front Oncol 2022;12:828505. [Crossref] [PubMed]
- Bercovici N, Guérin MV, Trautmann A, et al. The Remarkable Plasticity of Macrophages: A Chance to Fight Cancer. Front Immunol 2019;10:1563. [Crossref] [PubMed]
- Tetikkurt S, Taş F, Emre F, et al. Significant Neutrophilic Emperipolesis in Squamous Cell Carcinoma. Case Rep Oncol Med 2018;2018:1301562. [Crossref] [PubMed]
- Erfani N, Mehrabadi SM, Ghayumi MA, et al. Increase of regulatory T cells in metastatic stage and CTLA-4 over expression in lymphocytes of patients with non-small cell lung cancer (NSCLC). Lung Cancer 2012;77:306-11. [Crossref] [PubMed]
- Ferreira C, Barros L, Baptista M, et al. Type 1 Treg cells promote the generation of CD8+ tissue-resident memory T cells. Nat Immunol 2020;21:766-76. [Crossref] [PubMed]
- Zhou F, Qiao M, Zhou C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell Mol Immunol 2021;18:279-93. [Crossref] [PubMed]
- Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell 2017;170:1062-78. [Crossref] [PubMed]
- Wang X, Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget 2017;8:624-43. [Crossref] [PubMed]
- Blagih J, Buck MD, Vousden KH. p53, cancer and the immune response. J Cell Sci 2020;133:jcs237453. [Crossref] [PubMed]
- Song X, Chen Q, Wang J, et al. Clinical and prognostic implications of an immune-related risk model based on TP53 status in lung adenocarcinoma. J Cell Mol Med 2022;26:436-48. [Crossref] [PubMed]


