
The prognostic value and mechanisms of centromere protein M in patients with lung adenocarcinoma
Introduction
Lung cancer is one of the most common malignancies worldwide, inducing about 1.4 million global deaths every year (1,2), non-small cell lung cancer (NSCLC) accounts for about 85% of all lung cancers and can be divided into two histological subtypes, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (3,4). To date, emerging studies focused on identifying the underlying mechanisms of NSCLC. However, the specific mechanisms of NSCLC were still obscure, and no standard prognostic marker has been established for NSCLC, especially LUAD (5).
Previous studies showed that aberrant function of proteins involved in chromosome separation could induce aneuploidy which was found in many types of cancer (6,7). Aneuploidy is usually caused by an abnormal number and size of the centrosome, may accelerate tumorigenesis and carcinogenesis by causing chromosomes to separate unequally during mitosis (8-10). Recent researches indicated that centromere protein M (CENPM), also referred as proliferation associated nuclear element 1 (PANE1), is a kind of kinetochores protein which was detected in mouse mammary epithelium (11) and participated in affecting chromosome separation in the progress of cellular division (12). Together with several other centromere proteins, including CENPA, CENPC, CENPI and CENPH (13-15), which were homogeneous and closely correlated with tumors, manifested that high-expression of centromere protein family could have a significant impact on the proliferation and invasion of tumors (16,17).
To date, CENPM has been reported as a novel biomarker of hepatocellular carcinoma (18-20), pancreatic cancer (21), melanoma (22) and bladder cancer (23). However, the function of CENPM has not been identified in lung cancer. In the present study, we assessed the expression of CENPM in The Cancer Genome Atlas (TCGA) database and found that CENPM expression was significantly upregulated in both LUAD and LUSC tissues compared with normal tissues, and CENPM upregulation was relevant to the overall survival (OS) in LUAD patients. Thus, we discovered that CENPM was overexpressed in LUAD tissues and CENPM upregulation was closely concerned with higher pathological stages and poor prognosis in our patients. Additionally, we detected that CENPM inhibited cell apoptosis and affected cell cycle progression in vivo and in vitro via affecting the AKT1/mTOR signaling pathway. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-491/rc).
Methods
Bioinformatics analyses
The gene expression data were downloaded from the TCGA online database (https://genomecancer.ucsc.edu/). Initial validation was performed using gene expression data derived from normal tissues from the Genotype-Tissue Expression (GTEx) data combined with TCGA datasets. The gene expression of CENPM in LUAD and LUSC cohort were compared with normal tissue by independent sample t-test and paired sample t-test. Kaplan-Meier survival curves were used to compare the OS and disease specific survival (DSS) between LUAD and LUSC patients according to the CENPM gene expression. P<0.05 indicated statistical difference.
Gene Ontology (GO) analyses were used for the exploration of functional roles of CENPM, while Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used to classify the pathways in which CENPM might function. GO and KEGG analyses were conducted using the Database for Annotation Visualization and Integrated Discovery (DAVID) tool (http://david.ncifcrf.gov/). A false discovery rate (FDR) <0.05 in both GO and KEGG analyses was set as the threshold for significant enrichment.
Human tissues collections
Patients diagnosed with lung cancer in Fudan University affiliated Huadong Hospital from May 1, 2014 to Jun 30, 2015 were enrolled. This part of research was approved by Ethics Committee of Huadong Hospital. Enrollment criteria included (I) age between 18–80 years old; (II) received operation of lung cancer resection. Exclusion criteria included (I) pregnant or lactating; (II) lacked relevant clinical data. The tumor tissues and paired adjacent non-tumor (ANT) tissues were collected during the surgery. Medical records were collected, including age, gender, smoking history, tumor-node-metastasis (TNM) stage, receiving chemotherapy, postoperative recurrence and survival status. By Feb 1, 2021, we reviewed the medical records and pathological tests of 78 LUAD patients with paired tissues samples who had regularly postoperative follow-up. The study was approved by the ethics board of Huadong Hospital of Fudan University (No. 2021K010). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients.
Cell culture
LUAD cell lines (H1975, H292, H358 and A549) and human bronchial epithelial cells (16HBE) were used to perform the experiments. All cell lines were purchased from the Chinese Academy of Sciences Cell Bank (China). DMEM with 4.5 g/L glucose supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin were used to culture cells. Cells were incubated in a standard atmosphere with 5% CO2 and at a temperature of 37 ℃.
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cells or tissues were isolated using Trizol reagent (Invitrogen, USA) according to the RNA isolation kit instructions. RNA samples were reverse transcribed to cDNA, and RT-qPCR were then performed with ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA). GAPDH was regarded as an internal control. The results were presented with CT values and analyzed by the ΔΔCq method. Primer sequences for CENPM: forward 5'-GGCTGTGATGTCGGTGTTG-3' and reverse 5'-CAGGGACTCCTCTGTGTTCTG-3'.
Protein extraction and western blotting analysis
Protein samples were extracted using RIPA lysis buffer (Sigma-Aldrich, USA) with protease inhibitors and phosphatase inhibitors (Sigma-Aldrich, USA). Equal amounts of protein were separated on the 8% or 15% SDS-PAGE and transferred to a polyacrylamide gel electrophoresis (PVDF) membrane. Sodium dodecyl sulfate (SDS)-PAGE was used to separate proteins according to their molecular weight. Protein with higher molecular weight was separated with lower concentration gel and vice versa. After being blocked, the membranes were incubated with primary antibody at 4 ℃ for overnight. The primary antibodies were used as follows: anti-CENPM antibody (1:500 dilution; catalog Ab243820, Abcam), anti-AKT antibody (1:1,000 dilution; catalog 10176-2-AP, Proteintech), anti-p-AKT antibody (1:500 dilution; catalog 66444-1-Ig, Proteintech), anti-mTOR antibody (1:1,000 dilution; catalog Ab32028, Abcam), anti-p-mTOR antibody (1:500 dilution; catalog Ab109268, Abcam), and anti-GAPDH antibody (1:1,000 dilution; catalog 60004-1-1G, Proteintech). Horseradish peroxidase (HRP)-conjugated secondary antibodies were incubated for 80 min at room temperature. Protein bands were detected using chemiluminescence reagents (Beyotime Institute of Biotechnology, Shanghai, China).
Immunohistochemistry
Surgically resected tumor samples were fixed in formaldehyde and embedded in paraffin. The tissue blocks were cut into 5-µm sections that were collected onto slides. Paraffin-embedded sections were deparaffinized, rehydrated, and treated for 10 min with boiling citrate antigen-retrieval buffer and then incubated with 3% H2O2 for 10 min in a wet box. Tissue sections were incubated with primary antibodies against CENPM (1:800 dilution; Ab243820, Abcam), then incubated with a secondary antibody at room temperature for 20 min. Then stained the slides with 3,3'-diaminobenzidine (DAB) and counterstained with haematoxylin following the manufacturer’s protocol.
The CENPM expression level was calculated using the semi-quantitative rating system. The intensity of the stain in cytoplasm was score as follows: “negative” =0; “weak” =1; “moderate” =2 and “strong” =3. The stained cancer cell percentage was calculated based on the following method: 0 points (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), and 4 (≥76%). The final scores were calculated by multiplying the above two scores, resulting an overall score which range from 0 to 12. An immunostaining score ≤3 was classified as low CENPM expression and a score >3 was considered as high CENPM expression.
Construction of the lentivirus and cell transfection
To knockdown CENPM, three short hairpin RNA (shRNA) sequences (shCENPM-1, shCENPM-2 and shCENPM-3) were designed by the BLOCK-iTTM RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/). Lentiviral shRNA constructs were generated by inserting shRNA targeting sequences into pLKO.1 vector (Addgen, USA). A nonsense scrambled (SCR) shRNA sequence was used as a negative control (NC).
The shRNA sequences targeting CENPM were as follows:
- shCENPM-1: forward, 5'-CCGGTGAATTGACCTGATCGTGTTCTCGAGAACACGATCAGGTCAATTCTTTTTG-3'; reverse, 5'-AATTCAAAAAGAATTGACCTGATCGTGTTCTCGAGAACACGATCAGGTCAATTCA-3';
- shCENPM-2: forward, 5'-CCGGTCCTGATCGTGTTTGTGGTTCTCGAGAACCACAAACACGATCAGGTTTTTG-3'; reverse, 5'-AATTCAAAAACCTGATCGTGTTTGTGGTTCTCGAGAACCACAAACACGATCAGGA-3';
- shCENPM-3: forward, 5'-CCGGTCTGGCCCACACCTATCAAACTCGAGTTTGATAGGTGTGGGCCAGTTTTTG-3'; reverse, 5'-AATTCAAAAACTGGCCCACACCTATCAAACTCGAGTTTGATAGGTGTGGGCCAGA-3'.
The coding DNA sequence (CDS) region of CENPM (NM_024053.5), synthesized by GENEWIZ Company (Shanghai, China), was inserted into EcoR I/BamH I restriction sites of a pLVX-Puro vector. The synthesized core plasmid was confirmed by DNA sequencing (Majorbio, Shanghai). The primers used were as follows: CENPM-F, 5'-CGGAATTCATGTCGGTGTTGAGGCCC-3' (EcoR I); CENPM-R, 5'-CGGGATCCTCACAGGTCCTCCAGGGAGG-3' (BamH I). Cells transfected with pLVX-puro-CENPM served as the CENPM-OE group; cells transfected with pLVX-puro served as the vector group.
Subsequently, the above plasmids were respectively co-transfected with psPAX2 and pMD2.G (Addgen, USA) viral packaging plasmids into 293T cells (ATCC) by Lipofectamine 2000 (Invitrogen: Thermo Fisher Scientific, Inc.). The recombined lentiviral vectors were collected 48 h after transfection and used to infect H1975 and H292 cells. After infection for 48 h, 1.5 µg/mL puromycin was used continuously to screen and expand the cells. Stably-expressed cells were selected for subsequent experiments.
Cell proliferation assay
Cell proliferation assays were detected at 0, 24, 48 and 72 h. Approximately 5,000 cells/well of H1975 and H292 cells were seeded into 96-well plates according to the Cell Counting Kit-8 (CCK-8, SAB, CP002) protocol. Incubation lasted for 2 h at 37 ℃. Absorbance was detected at 450 nm using Infinite 200 Pro microplate reader.
Flow cytometry analysis of cell cycle and apoptosis
DNA content was evaluated by flow cytometry (FACS). H1975 and H292 cells were harvested washed twice with cold PBS, then fixed in 70% cold ethanol and stored at 4 ℃ for 12 h. Then, the cells were treated with RNase A and were stained with 50 µg/mL propidium iodide (PI) (C1052, Beyotime Biotechnology, China) for 30 min at 37 ℃ in the dark. The CytoFLEX flow cytometers (Beckman Coulter, China) was used to measure DNA content and the data were analyzed using the FlowJo software.
Apoptosis analysis by Annexin V fluorescein isothiocyanate (FITC)/PI double staining
Annexin V and PI staining were carried out using the Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology, China). The transfected cells were washed with ice-cold PBS and resuspended in binding buffer at a concentration of 106 cells/mL. The experimental procedure refers to the instructions of product manual: add 5 µL of FITC-labeled Annexin V and 10 µL of PI and mixed, incubated at room temperature in the dark for 15 min. The stained cells were then analyzed by flow cytometry. The data was analyzed by FlowJo software.
Cell migration and invasion assays
Approximately 5×104 lentivirus-infected H1975 and H292 cells were seeded into Transwell chambers with pore size inserts of 8 µm (catalog 3422, Corning, USA). Matrigel (catalog 3422, Corning, USA) was diluted with DMEM followed by incubation in the chambers overnight. The cells were cultured in the upper chambers and were then allowed to invade for 24 h. All of the lower chambers were filled with DMEM supplemented with 20% FBS. Upon completion of migration or invasion, the chambers were fixed with 4% paraformaldehyde at 4 ℃ for 20 min and stained with 0.5% crystal violet at 37 ℃ for 20 min carefully. Images were observed by light microscope and analyzed to calculate the Transwell capacity.
In vivo tumor xenograft study
Twelve male BALB/c nude mice at 6 weeks of age, weighing 18–22 g, were obtain from Sipul-Bikai Experimental Animal Co., Ltd. (Shanghai, China). The animal experimental procedures of the study followed the guidelines of the Huadong Hospital of Fudan University Animal Ethics Committee and were approved by the Animal Care and Use Committee. Mice were divided randomly into shNC and shCENPM groups, with six mice in each group. H1975 cells (100 µL, 5×106) stably transfected with shCEPNM or shNC were harvested and inoculated subcutaneously in the mice. Two weeks after injection, the size of the tumor was measured by caliper every 3 days. The tumor volume was calculated by the following formula: volume =0.5 × length × width2. Thirty-three days after the injection, the tumors were removed and weighed successively. The animal experiments were performed under a project license (No. 20210010) granted by the ethics board of Huadong Hospital of Fudan University, in compliance with the Guidelines and Suggestions for Laboratory Animals (Ministry of Science and Technology of the People’s Republic of China).
TUNEL staining
The TUNEL staining was used to evaluate cell apoptosis in tissue. In Situ Apoptosis Detection Kit was used with manufacturer’s instruction (Beyotime Institute of Biotechnology, Shanghai, China). The nuclei were stained with DAPI (catalog C1005, Beyotime Institute of Biotechnology, Shanghai, China), and the fluorescent microscopy was used to evaluate cell apoptosis.
Immunofluorescence staining
The tissues were stored in 4% paraformaldehyde and sectioned at 30 µm, and immunofluorescence staining for Ki67 (catalog 15580, Abcam) and DAPI were performed. The sections were incubated with Ki67 antibodies overnight at 4 ℃. Anti-rabbit secondary antibodies were added and incubated for 2 h at 37 ℃, and sections were then washed three times with phosphate buffered saline with Tween (PBST). After final washing, sections were protected with coverslips, with the nucleus visualized with DAPI. The tissues were observed and analyzed using fluorescence microscope.
Statistical analysis
Data are presented as the mean values ± standard deviation (SD) and were performed using t-tests or one-way ANOVA. P values <0.05 were considered to indicate a statistically significant difference. Statistical analyses were achieved using SPSS 23.0 software (IBM, USA) and GraphPad Prism version 7 (GraphPad Software, USA).
Results
CENPM is upregulated in lung cancer and associated with the prognosis of LUAD patients via bioinformatics analysis
The data extracted from the TIMER database (http://timer.cistrome.org/) revealed that CENPM expression was highly expressed in almost all tumor types, including LUAD and LUSC (Figure 1A). TCGA database revealed that centromere protein family members (CENPM, CENPA, CENPF, CENPU, CENPO and CENPH) were significantly upregulated in patients with LUAD and LUSC (P<0.001) (Figure 1B, Figure S1A). Independent and paired analysis showed that CENPM was sharply increased in LUAD and LUSC patients compared with healthy controls (P<0.001) (Figure 1C,1D, Figure S1B,S1C). Upregulation of CENPM in both LUAD and LUSC patients showed good prediction effect with the area under the curve (AUC) of 0.847 and 0.968 respectively (Figure 1E, Figure S1D). OS and DSS data extracted from TCGA database revealed that LUAD patients with high CENPM expression presented decreased survival rates (P=0.001 and P<0.001), while LUSC patients showed poor relationship between high CENPM expression with OS or DSS (P>0.05) (Figure 1F, Figure S1E). We also analyzed the data extracted from Kaplan-Meier Plotter database (http://kmplot.com/analysis/index) and found the similar trends (Figure 1G).
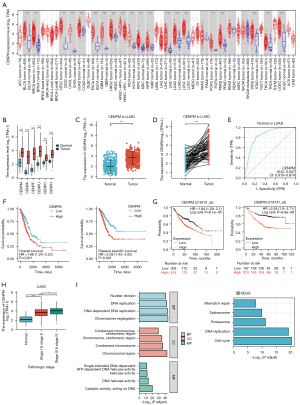
To explore the relationship between CENPM expression with different clinical pathological stages, we found that the higher CENPM mRNA levels, the higher LUAD clinical pathological stages in TCGA database (Figure 1H). The major GO functional and top KEGG pathways of CENPM were illustrated (Figure 1I, Table 1). The top significantly enriched GO domains were associated with chromosome segregation, chromosomal region and catalytic activity acting on DNA, respectively. The top significant KEGG pathways of CENPM were enriched in cell cycle, DNA replication, proteasome, spliceosome and mismatch repair. All these bioinformatics results indicated that CENPM was upregulated in lung cancer and associated with the prognosis of LUAD patients.
Table 1
| Ontology | ID | Description | Gene ratio | Bg ratio | P value | P adjust | q value |
|---|---|---|---|---|---|---|---|
| BP | GO:0007059 | Chromosome segregation | 84/669 | 321/18,670 | 6.97e−49 | 2.47e−45 | 2.00e−45 |
| BP | GO:0006261 | DNA-dependent DNA replication | 61/669 | 153/18,670 | 6.64e−48 | 1.18e−44 | 9.52e−45 |
| BP | GO:0006260 | DNA replication | 76/669 | 274/18,670 | 3.14e−46 | 3.71e−43 | 3.00e−43 |
| BP | GO:0000280 | Nuclear division | 89/669 | 407/18,670 | 8.17e−45 | 7.23e−42 | 5.85e−42 |
| CC | GO:0098687 | Chromosomal region | 84/690 | 349/19,717 | 1.75e−46 | 8.84e−44 | 5.78e−44 |
| CC | GO:0000793 | Condensed chromosome | 63/690 | 223/19,717 | 1.78e−39 | 4.50e−37 | 2.94e−37 |
| CC | GO:0000775 | Chromosome, centromeric region | 58/690 | 193/19,717 | 4.63e−38 | 7.80e−36 | 5.10e−36 |
| CC | GO:0000779 | Condensed chromosome, centromeric region | 43/690 | 118/19,717 | 1.83e−32 | 2.31e−30 | 1.51e−30 |
| MF | GO:0140097 | Catalytic activity, acting on DNA | 48/673 | 213/17,697 | 5.82e−24 | 3.62e−21 | 3.06e−21 |
| MF | GO:0003678 | DNA helicase activity | 26/673 | 81/17,697 | 1.19e−17 | 3.70e−15 | 3.12e−15 |
| MF | GO:0004386 | Helicase activity | 33/673 | 163/17,697 | 2.18e−15 | 4.53e−13 | 3.82e−13 |
| MF | GO:0017116 | Single-stranded DNA-dependent ATP-dependent DNA helicase activity | 13/673 | 20/17,697 | 1.88e−14 | 2.35e−12 | 1.98e−12 |
| KEGG | hsa04110 | Cell cycle | 38/332 | 124/8,076 | 1.33e−23 | 2.88e−21 | 2.47e−21 |
| KEGG | hsa03030 | DNA replication | 22/332 | 36/8,076 | 3.64e−22 | 3.93e−20 | 3.37e−20 |
| KEGG | hsa03050 | Proteasome | 18/332 | 46/8,076 | 7.05e−14 | 5.07e−12 | 4.35e−12 |
| KEGG | hsa03040 | Spliceosome | 29/332 | 151/8,076 | 2.00e−12 | 1.08e−10 | 9.25e−11 |
| KEGG | hsa03430 | Mismatch repair | 11/332 | 23/8,076 | 4.18e−10 | 1.66e−08 | 1.43e−08 |
CENPM, centromere protein M; Bg ratio, background ratio; BP, biological process; CC, cellular component; MF, molecular function; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Upregulation of CENPM in LUAD patients correlates with higher pathological stages and poor survival rates
To determine the effect of CENPM on LUAD patients, we collected the data from 78 LUAD patients who received the lung surgery in our hospital. The basic characteristics were listed (Table 2). Western blotting analysis was conducted with 20 matched pairs of LUAD tissues and ANT tissues. We found that CENPM expression was increased in LUAD tissue samples compared with ANT (P<0.0001) (Figure 2A). All of the 78 paired samples were tested by RT-qPCR, and revealed the CENPM mRNA upregulation in LUAD tissues (P<0.01) (Figure 2B). Immunohistochemical analysis indicated that the intensity of the stain in cytoplasm were significantly stronger in higher pathological stages compared with lower stages (Figure 2C). Moreover, CENPM mRNA levels increased with the clinical pathological stages (P<0.05) (Figure 2D). During follow-up studies, LUAD patients with high CENPM expression presented decreased survival rates (P<0.05) (Figure 2E). Bioinformatics-based analysis from TCGA and Kaplan-Meier Plotter database also confirmed that increased CENPM was correlated with higher TNM and pathological stages (P<0.05) (Figure 1H, Figure S2A-S2D), and poor OS was closely related to higher CENPM in different pathological stages (P<0.05) (Figure S2E-S2G).
Table 2
| Characteristic | Low expression of CENPM | High expression of CENPM | P | χ2 |
|---|---|---|---|---|
| N | 39 | 39 | ||
| T stage, n (%) | 0.002 | 13.84 | ||
| T1 | 16 (20.51) | 9 (11.54) | ||
| T2 | 19 (24.36) | 24 (30.77) | ||
| T3 | 3 (3.85) | 4 (5.13) | ||
| T4 | 1 (1.28) | 2 (2.56) | ||
| N stage, n (%) | <0.001 | – | ||
| N0 | 29 (37.18) | 23 (29.49) | ||
| N1 | 5 (6.41) | 9 (11.54) | ||
| N2 | 5 (6.41) | 7 (8.97) | ||
| Pathologic stage, n (%) | <0.001 | 11.71 | ||
| Stage I | 26 (33.33) | 18 (23.08) | ||
| Stage II | 8 (10.26) | 12 (15.38) | ||
| Stage III | 5 (6.41) | 9 (11.54) | ||
| Gender, n (%) | 0.017 | 4.70 | ||
| Female | 24 (30.77) | 19 (24.36) | ||
| Male | 15 (19.23) | 20 (25.64) | ||
| Age, n (%) | 0.017 | 5.65 | ||
| ≤65 years | 17 (21.79) | 21 (26.92) | ||
| >65 years | 22 (28.21) | 18 (23.08) | ||
| Smoker, n (%) | 1.000 | – | ||
| No | 6 (7.69) | 5 (6.41) | ||
| Yes | 33 (42.31) | 34 (43.59) | ||
| Number pack years smoked, n (%) | 0.006 | 5.61 | ||
| <40 | 19 (24.36) | 17 (21.80) | ||
| ≥40 | 20 (25.64) | 22 (28.20) | ||
| Histology, n (%) | 0.640 | – | ||
| Squamous cell carcinoma | 5 (6.41) | 7 (8.97) | ||
| Adenocarcinoma | 30 (38.46) | 36 (46.15) | ||
| Age, median [IQR], years | 67 [60, 73] | 63 [57, 71] |
CENPM, centromere protein M; T, tumor; N, node; IQR, interquartile range.
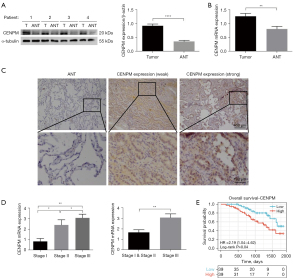
We further analyzed the association between CENPM expression and the clinical pathological features in our patients. As shown in Table 2, strong associations were observed between CENPM expression and pathological stage (P<0.001), T classification (P=0.002) and N classification (P<0.001). We also analyzed the relative risks indicated by CENPM in the prognosis of LUAD. Univariate and multivariate Cox analysis confirmed that poor OS was concerned with higher CENPM expression and higher TNM stages (Table 3). Taken together, these results indicated that upregulation of CENPM in LUAD patients was associated with higher pathological stages and poor survival rates.
Table 3
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |||
| Age | 1.224 (0.917–2.635) | 0.173 | – | – | ||
| Gender | 1.080 (0.803–2.426) | 0.644 | – | – | ||
| T stage | 2.317 (1.591–3.735) | <0.001 | 1.564 (1.015–2.856) | 0.041 | ||
| N stage | 2.241 (1.631–3.986) | <0.001 | 1.526 (1.014–2.762) | 0.038 | ||
| Pathologic stage (TNM) | 2.821 (2.173–3.958) | <0.001 | 1.746 (1.231–3.833) | 0.003 | ||
| Smoker | 0.894 (0.592–2.348) | 0.591 | – | – | ||
| Number pack years smoked (≥40 vs. <40) | 1.079 (0.754–1.928) | 0.697 | – | – | ||
| CENPM (high vs. low) expression | 1.656 (1.242–4.232) | <0.001 | 1.796 (1.102–3.749) | 0.024 | ||
CENPM, centromere protein M; OS, overall survival; LUAD, lung adenocarcinoma; CI, confidence interval; T, tumor; N, node; TNM, tumor-node-metastasis.
CENPM promotes the proliferation in LUAD cells
To determine the expression of CENPM in LUAD cell lines, different LUAD cell lines (H1975, H292, H358 and A549) and human bronchial epithelial cells (16HBE) were evaluated. We found that CENPM was upregulated in all LUAD cell lines compared with 16HBE (P<0.05) (Figure 3A). Then, H1975 and H292 cell lines were selected for further study due to their relatively higher and lower expression levels respectively. Three effective shRNAs were transfected to interfere the expression of CENPM in H1975 cells, and the inhibiting effects were confirmed compared to shNC and control group (P<0.0001). Due to the knockdown effect, shCENPM-1 and shCENPM-2 were used in the following experiments (Figure 3B). The CENPM plasmids were transfected to H292 cells, and the overexpression effect was then confirmed (P<0.05) (Figure 3C). Thus, CENPM knockdown and overexpression cell lines were successfully constructed.
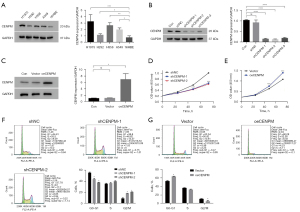
Time-dependent proliferation was evaluated by CCK-8 assay. LUAD cells transfected with shCENPM-1 and shCENPM-2 had significantly lower values compared with shNC group (P<0.001) (Figure 3D), while CENPM overexpression improved values compared with vector group (P<0.001) (Figure 3E). The results indicated that CENPM could promote the proliferation in LUAD cells.
CENPM affects the cell cycle in LUAD cells
To investigate the effect of CENPM on cell cycle, we performed the cytometric analysis. We found that down-regulation of CENPM contributed to decreased proportion of cells in G0/G1 phase, and increased in G2/M phase (P<0.05) (Figure 3F). Additionally, increased G0/G1 proportion cells and decreased G2/M proportion cells were witnessed after CENPM up-regulation in LUAD cells (P<0.05) (Figure 3G). These results indicated that CENPM affected the cell cycle and might be involved the process of cell mitosis in LUAD cells.
CENPM inhibits the cell apoptosis in LUAD cells
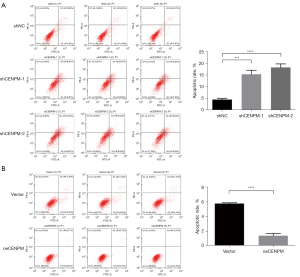
To better understand the role of CENPM in cell apoptosis, flow cytometric analysis by Annexin V/PI staining were then performed. We showed that the proportion rate of early and late apoptotic cells in shCENPM-1 and shCENPM-2 groups were sharply increased than those in shNC group (P<0.001) (Figure 4A), while CENPM overexpression significantly decreased compared with vector group (P<0.001) (Figure 4B). These results indicated that CENPM inhibited the cell apoptosis in LUAD cells.
CENPM promotes the migration and invasion in LUAD cells
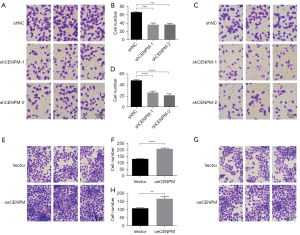
Transwell assays were performed to evaluate the migration and invasion function of CENPM in LUAD cells. Cells with migration capacity can move from the upper chambers without FBS to the lower chambers with a high concentration of FBS. Our results showed that down-regulation of CENPM could efficiently hinder the migration (P<0.001) (Figure 5A,5B) and invasion (P<0.0001) (Figure 5C,5D) capacity compared with shNC group. While upregulated CENPM could significantly enhance the migration (P<0.0001) (Figure 5E,5F) and invasion (P<0.01) (Figure 5G,5H) capacity in LUAD cells. These results indicated that CENPM promoted the migration and invasion in LUAD cells.
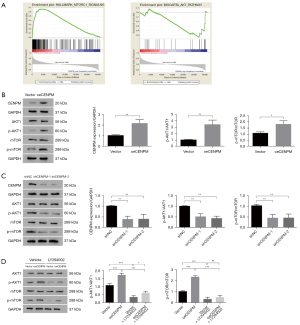
CENPM function could be regulated via AKT1/mTOR signaling pathway
In an attempt to determine which pathway may be involved in CENPM-mediated LUAD progression, we performed analysis in Gene Set Enrichment Analyses (GSEA) in the published TCGA-LUAD database (n=517). We found the enrichment plot results showed that mTOR complex 1 (mTORC1) and AKT were significantly related to the function of CENPM (Figure 6A). Thus, we focused on the relationship between CENPM function in AKT1/mTOR signaling pathway. We found that CENPM overexpression led to AKT1/mTOR pathway activation, as evidenced by increased p-AKT1/AKT1 ratio (P<0.01) and p-mTOR/mTOR ratio (P<0.05) in LUAD cells (Figure 6B). Moreover, AKT1/mTOR activity was declined after CENPM down-regulation (Figure 6C) or pretreated with LY294002 in LUAD cells (Figure 6D).
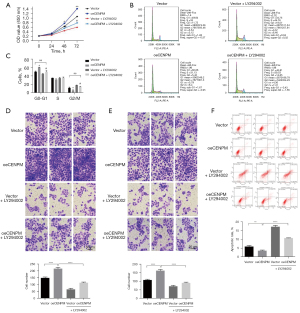
Furthermore, we evaluated the cell proliferation, cell cycle, cell apoptotic rate, cell migration and invasion in CENPM overexpression (oeCENPM) LUAD cells with or without the treatment of LY294002. The results showed that LY294002 reversed the biological function of CENPM overexpression, i.e., affecting cell proliferation (P<0.05) (Figure 7A), cell cycle (P<0.01) (Figure 7B,7C), cell migration and invasion capacity (P<0.01) (Figure 7D,7E), and apoptosis (P<0.01) (Figure 7F) compared with oeCENPM treatment. These results indicated that upregulation of CENPM enhanced the malignant degree of LUAD cells through AKT1/mTOR signaling pathway.
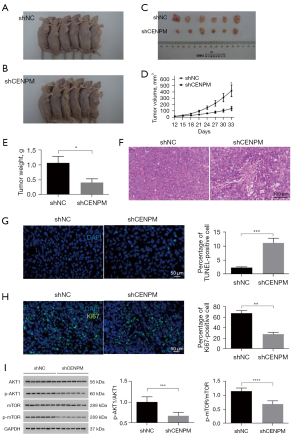
CENPM knockdown inhibits cell proliferation and promotes cell apoptosis in vivo
In vivo, we conducted subcutaneous xenograft experiments in nude mice, H1975 cells with shCENPM and shNC were injected into the armpit of male nude mice. We found that downregulated CENPM repressed tumor growth 33 days after injection (P<0.05) (Figure 8A-8E). We also found that disorganized structure in the xenograft of shCENPM group (Figure 8F), and increased TUNEL positive cells (P<0.001) (Figure 8G) as well as decreased positivity for Ki67 (P<0.01) (Figure 8H) in shCENPM group. Moreover, we showed that AKT1/mTOR activity were significantly declined in mice injected with shCENPM H1975 cells (P<0.001/P<0.0001) (Figure 8I), which clearly indicated that CENPM could promote tumor growth through AKT1/mTOR signaling pathway in LUAD.
Discussion
Nowadays, diagnostic and treatment technologies for LUAD patients have continuously developed. In particular, clinical application of novel targeted medicines has effectively improved the survival rate of LUAD patients (24). However, the prognosis for advanced LUAD patients remains dismal, and the 5-year survival rate is still lower than 20% (25). Thus, searching for the new therapeutic target is a current research focal point. Kjeldsen. pointed out that chromosome instability, the process that gives rise to aneuploidy, could promote tumorigenesis by increasing genetic heterogeneity and promoting tumor evolution (26). Recent studies suggested that aneuploidy might have clinical relevance as a prognostic marker and potential therapeutic target in tumors (6,27).
CENPM is a component of the CENPA-NAC (nucleosome-associated) complex, and plays a critical role in assembly of kinetochores proteins, mitotic progression and chromosome segregation during cell division (28). Recent published studies showed that CENPM exerted its pro-tumorigenic function by regulating cell cycle-associated protein expression via p53 signaling pathway (18) and mTOR/p70S6K signaling pathway (21), which indicated that CENPM could be used as a prognostic marker in tumor diagnosis (11). However, the mechanism between CENPM and lung cancer has not been explored. Our study determined the function of CENPM in the proliferation, cell cycle, metastasis and apoptosis of LUAD.
Our study revealed that CENPM was upregulated in both LUAD and LUSC compared with healthy controls. However, CENPM upregulation was only relevant to the OS in LUAD patients, indicated that CENPM played a key role in the prognostic in the LUAD process. Then, we found that CENPM could promote cell proliferation, cause cell cycle arrest at G0/G1 phase, enhance cell migration and invasion capacity, promote apoptosis in LUAD cell lines and promote the growth of xenografts in nude mice. These altered biological function were correlated with the regulation of p-AKT1 and p-mTOR expression, which indicated that AKT1/mTOR signaling pathway acted as underlying mechanism of CENPM.
The PI3K/AKT/mTOR pathway played important role in regulating cell proliferation, growth, cell size, metabolism and motility (29). In NSCLC, the PI3K/AKT/mTOR pathway has been heavily implicated in both tumorigenesis and the progression of disease (30,31). Previous studies have shown that the expression of PI3K, AKT and mTOR upregulated in NSCLC (31,32). The preclinical studies showed that AKT activation attributed to loss of PTEN, EGFR, PIK3CA mutation or HER2 amplification in NSCLC cell lines (29,33). Upregulation of the mTOR pathway has also been illustrated in significant proportions of NSCLC tumors, with increased p-mTOR in up to 90% of patients with LUAD, 60% of patients with large cell carcinoma and 40% of patients with LUSC (34). The downstream products of mTOR activation, S6K and 4E-BP1 have also been identified in up to 58% and 25% of NSCLC specimens, respectively (35). PI3K, AKT and mTOR and dual mTORC1 and mTOR complex 2 (mTORC2) inhibitors exhibit promise in clinical trials but are to be approved yet for human cancer therapy (36). However, some mutations (such as EGFR in H1975 cell lines) in LUAD cell lines may contribute to the activation of PI3K/AKT/mTOR pathway. Our results found different activity of AKT/mTOR pathway between shCENPM group and shNC group, which indicated that the CENPM-mediated AKT/mTOR activation was distinguished from EGFR mutation. In our study, CENPM may regulate the activity of the kinase by interacting with the kinase domain and play an important role in p-AKT and p-mTOR. However, the specific molecular mechanisms remain unclear and will be explored in future studies.
Conclusions
To sum up, we revealed upregulated expression of CENPM expression in human LUAD tissues relative to normal lung tissues, and upregulated CENPM also predicts a poor prognosis in LUAD patients and is associated with higher pathological stages. CENPM could affect cell proliferation, cell cycle, cell migration and invasion capacity, and apoptosis in LUAD cell lines via regulation of AKT1/mTOR signaling pathway.
Our findings identify a potential role for the CENPM gene as a prognostic biomarker and a new therapeutic target in LUAD by activating the AKT1/mTOR signaling pathway. Notably, more investigations in animal research and clinical practice are required to obtain more complete elucidation.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-491/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-491/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-491/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-491/coif). All authors report funding from National Key R&D Program for Intergovernmental International Science and Technology Innovation Cooperation Key Project (grant No. 2019YFE0105600). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethics board of Huadong Hospital of Fudan University (No. 2021K010). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients. The animal experiments were performed under a project license (No. 20210010) granted by the ethics board of Huadong Hospital of Fudan University, in compliance with the Guidelines and Suggestions for Laboratory Animals (Ministry of Science and Technology of the People’s Republic of China).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thandra KC, Barsouk A, Saginala K, et al. Epidemiology of lung cancer. Contemp Oncol (Pozn) 2021;25:45-52. [Crossref] [PubMed]
- Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol 2021;33:40-6. [Crossref] [PubMed]
- Relli V, Trerotola M, Guerra E, et al. Abandoning the Notion of Non-Small Cell Lung Cancer. Trends Mol Med 2019;25:585-94. [Crossref] [PubMed]
- Mithoowani H, Febbraro M. Non-Small-Cell Lung Cancer in 2022: A Review for General Practitioners in Oncology. Curr Oncol 2022;29:1828-39. [Crossref] [PubMed]
- Zhang Y, Luo G, Etxeberria J, et al. Global Patterns and Trends in Lung Cancer Incidence: A Population-Based Study. J Thorac Oncol 2021;16:933-44. [Crossref] [PubMed]
- Chiarle R. Solving the chromosome puzzle of aneuploidy in cancer. Genes Dev 2021;35:1073-5. [Crossref] [PubMed]
- Golas MM, Gunawan B, Cakir M, et al. Evolutionary patterns of chromosomal instability and mismatch repair deficiency in proximal and distal colorectal cancer. Colorectal Dis 2022;24:157-76. [Crossref] [PubMed]
- Li R, Zhu J. Effects of aneuploidy on cell behaviour and function. Nat Rev Mol Cell Biol 2022;23:250-65. [Crossref] [PubMed]
- Simonetti G, Bruno S, Padella A, et al. Aneuploidy: Cancer strength or vulnerability? Int J Cancer 2019;144:8-25. [Crossref] [PubMed]
- Garribba L, Santaguida S. The Dynamic Instability of the Aneuploid Genome. Front Cell Dev Biol 2022;10:838928. [Crossref] [PubMed]
- Liu Y, Yu W, Ren P, et al. Upregulation of centromere protein M promotes tumorigenesis: A potential predictive target for cancer in humans. Mol Med Rep 2020;22:3922-34. [Crossref] [PubMed]
- Vicars H, Karg T, Warecki B, et al. Kinetochore-independent mechanisms of sister chromosome separation. PLoS Genet 2021;17:e1009304. [Crossref] [PubMed]
- Mahlke MA, Nechemia-Arbely Y. Guarding the Genome: CENP-A-Chromatin in Health and Cancer. Genes (Basel) 2020;11:810. [Crossref] [PubMed]
- Fellmeth JE, McKim KS. Meiotic CENP-C is a shepherd: bridging the space between the centromere and the kinetochore in time and space. Essays Biochem 2020;64:251-61. [Crossref] [PubMed]
- Hu L, Huang H, Hei M, et al. Structural analysis of fungal CENP-H/I/K homologs reveals a conserved assembly mechanism underlying proper chromosome alignment. Nucleic Acids Res 2019;47:468-79. [Crossref] [PubMed]
- Anjur-Dietrich MI, Kelleher CP, Needleman DJ. Mechanical Mechanisms of Chromosome Segregation. Cells 2021;10:465. [Crossref] [PubMed]
- Kim T. Epigenetic control of centromere: what can we learn from neocentromere? Genes Genomics 2022;44:317-25. [Crossref] [PubMed]
- Xiao Y, Najeeb RM, Ma D, et al. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J Exp Clin Cancer Res 2019;38:458. [Crossref] [PubMed]
- Zou Y, Sun Z, Sun S. LncRNA HCG18 contributes to the progression of hepatocellular carcinoma via miR-214-3p/CENPM axis. J Biochem 2020;168:535-46. [Crossref] [PubMed]
- Ren H, Wei ZC, Sun YX, et al. ATF2-Induced Overexpression of lncRNA LINC00882, as a Novel Therapeutic Target, Accelerates Hepatocellular Carcinoma Progression via Sponging miR-214-3p to Upregulate CENPM. Front Oncol 2021;11:714264. [Crossref] [PubMed]
- Zheng C, Zhang T, Li D, et al. Upregulation of CENPM facilitates tumor metastasis via the mTOR/p70S6K signaling pathway in pancreatic cancer. Oncol Rep 2020;44:1003-12. [Crossref] [PubMed]
- Chen J, Wu F, Shi Y, et al. Identification of key candidate genes involved in melanoma metastasis. Mol Med Rep 2019;20:903-14. [Crossref] [PubMed]
- Kim WT, Seo SP, Byun YJ, et al. The Anticancer Effects of Garlic Extracts on Bladder Cancer Compared to Cisplatin: A Common Mechanism of Action via Centromere Protein M. Am J Chin Med 2018;46:689-705. [Crossref] [PubMed]
- Xu JY, Zhang C, Wang X, et al. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell 2020;182:245-261.e17. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Kjeldsen E. Congenital Aneuploidy in Klinefelter Syndrome with B-Cell Acute Lymphoblastic Leukemia Might Be Associated with Chromosomal Instability and Reduced Telomere Length. Cancers (Basel) 2022;14:2316. [Crossref] [PubMed]
- Lukow DA, Sheltzer JM. Chromosomal instability and aneuploidy as causes of cancer drug resistance. Trends Cancer 2022;8:43-53. [Crossref] [PubMed]
- Liu X, Wang H, Zhao G. Centromere Protein A Goes Far Beyond the Centromere in Cancers. Mol Cancer Res 2022;20:3-10. [Crossref] [PubMed]
- Peng Y, Wang Y, Zhou C, et al. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front Oncol 2022;12:819128. [Crossref] [PubMed]
- Tan AC. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac Cancer 2020;11:511-8. [Crossref] [PubMed]
- Sanaei MJ, Razi S, Pourbagheri-Sigaroodi A, et al. The PI3K/Akt/mTOR pathway in lung cancer; oncogenic alterations, therapeutic opportunities, challenges, and a glance at the application of nanoparticles. Transl Oncol 2022;18:101364. [Crossref] [PubMed]
- Wang J, Zhou F, Li F, et al. Autocrined leptin promotes proliferation of non-small cell lung cancer (NSCLC) via PI3K/AKT and p53 pathways. Ann Transl Med 2021;9:568. [Crossref] [PubMed]
- Wu R, Yuan B, Li C, et al. A narrative review of advances in treatment and survival prognosis of HER2-positive malignant lung cancers. J Thorac Dis 2021;13:3708-20. [Crossref] [PubMed]
- Krencz I, Sebestyen A, Khoor A. mTOR in Lung Neoplasms. Pathol Oncol Res 2020;26:35-48. [Crossref] [PubMed]
- Jeong M, Jeong MH, Kim JE, et al. TCTP protein degradation by targeting mTORC1 and signaling through S6K, Akt, and Plk1 sensitizes lung cancer cells to DNA-damaging drugs. Sci Rep 2021;11:20812. [Crossref] [PubMed]
- Sun SY. mTOR-targeted cancer therapy: great target but disappointing clinical outcomes, why? Front Med 2021;15:221-31. [Crossref] [PubMed]


