
MAN2A1 predicts prognosis and progression through cancer-related pathways in colorectal cancer
Introduction
Colorectal cancer (CRC) is a common digestive system carcinoma, CRC is the third commonly diagnosed malignant tumor and the fourth malignancy leading to death (1). Approximately 44% of CRC patients were diagnosed with stage Ⅰ-Ⅱ, and the 5-year survival rate of stage Ⅰ and stage Ⅱ was 91% and 82% respectively (2). However, 35% of patients were diagnosed with metastatic disease and 50% of patients diagnosed with non-metastatic CRC ended up manifesting with metastatic disease, leading to low survival rates (3-5).
With the development of molecular biology and bioinformatics, differentially expressed genes (DEGs) between metastatic tumor and in situ tumor can be identified. These DEGs are relevant to the pathological stage, metastatic and prognosis of patients (6). The wide application of various bioinformatics tools also make efforts to predict the possible mechanisms of genes (7,8).
In this study, we screened colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) datasets from The Cancer Genome Atlas (TCGA) databases to identify DEGs between lymph node metastasis (LNM) patients and non-LNM patients which also associated with prognosis. In addition, we constructed a protein-protein interactive (PPI) network and used Cytoscape molecular complex detection (MCODE) plus to find hub genes. We explored these hub genes, found the association between Mannosidase alpha class 2A member 1 (MAN2A1) expression level and prognosis of CRC patients, and verified RNA expression level in CRC patients of our cohort. Therefore, we identified MAN2A1 as a candidate gene for biomarker related to diagnosis and prognosis of CRC patients. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-629/rc).
Methods
Tumor samples and ethics approval
The fresh tumor specimens and corresponding adjacent normal tissues were obtained from CRC patients who underwent surgery at Sir Run Run Shaw Hospital, affiliated to Zhejiang University, from 2018 to 2020. These tissues were stored at −80 ℃ at once after surgery (Table 1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Sir Run Run Shaw Hospital, affiliated to Zhejiang University approved all study protocols (approval ID. 20200619-34). Individual consent for this retrospective study was waived.
Table 1
| Variables | Data |
|---|---|
| Total number | 26 |
| Age (years), mean ± SD | 61.08±12.46 |
| Minimum | 37 |
| Maximum | 80 |
| Gender, n (%) | |
| Male | 13 (50.00) |
| Female | 13 (50.00) |
| Regional lymph node invasion, n (%) | |
| No | 10 (38.46) |
| Yes | 16 (61.54) |
SD, standard deviation.
Data acquisition
We downloaded CRC transcriptome profiles from TCGA database through the Genomic Data Commons (GDC) Portal. The public data included RNA-seq profiles of 272 CRC lymph node metastatic tissues (N1, N2) and 368 non-metastatic tissues (N0).
Identification of intersection genes
We merged RNA raw counts data by Python to analyze the RNA-seq data and converted gene ID to gene symbol (Homo sapiens). Then, batch processing of univariate Cox regression analysis was performed to identify genes associated with prognosis of CRC (P≤0.05). The DEGs were identified with edgeR package in Sangerbox tool (P≤0.05). Venn diagram was used to identify intersection genes of the above conditions.
PPI network
The interaction of the intersection genes were analyzed with Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org) (9). Then, Cytoscape (http://www.cytoscape.org/) software was used for establishing PPI network. In addition, Cytoscape MCODE plus was used to analyze modules and got hub genes (10).
Enrichment analysis
To identified the biological significance of the intersection genes, the KEGG Orthology Based Annotation System (KOBAS) 3.0 (11) and Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tools (12,13) were used for enrichment analysis. The DAVID was used to reveal Gene Ontology (GO) (P≤0.05), KOBAS 3.0 was used to reveal (Kyoto Encyclopedia of Genes and Genomes) KEGG pathways (corrected P value ≤0.05).
Detect MAN2A1 mRNA expression
Total RNA was isolated from the tissues using RNeasy Mini Kit (QIAGEN, Dusseldorf, Germany). The concentration and purity of RNA were detected with the NanoDrop 2000 spectrophotometer. Total RNA (1000 ng) was converted to complementary DNA (cDNA) by RevertAid First Strand cDNA Synthesis Kit (MA, USA, thermo scientific). We used LightCycler96 to conduct real-time polymerase chain reaction (RT-PCR) by SYBR Green (Nanjing, China, Vazyme). We used two-step amplification reaction as follows: 25 ℃ for 5 min for preincubation, then 42 ℃ for 60 min and 70 ℃ for 5 min. GAPDH expression was utilized as an endogenous control. Quantitative analysis was calculated by the 2−ΔCt method. Primers were designed as follows: MAN2A1 forward, 5'-AAGTCAGCGCAGTTTGGGAT-3' and reverse 5'-CCACAGACTGTCCTTCTATTCCC-3'.
Gene set enrichment analysis (GSEA)
GSEA is a computational mathematics method to interpret gene expression data and demonstrate differences between two biological group (14,15). GSEA 4.1.0 software was used to analyze the biological functions of MAN2A1 in CRC. CRC patients were divided into two groups according to MAN2A1 expression levels. We used the Molecular Signatures Database (16,17) (MsigDB, https://www.gseamsigdb.org/gsea/msigdb/) to analyze KEGG pathways and GO terms to explore the potential biological mechanism and function of MAN2A1, GSEA parameters were repeating the analysis 2000 times at a time and default weighted enrichment statistics. false discovery rate (FDR) q‐value ≤0.05 was set as cut-off criteria.
Statistical analysis
Sangerbox (http://vip.sangerbox.com/) is a comprehensive bioinformatics analysis tool that can carry out bioinformatics analysis and visualize results (18). Kaplan-Meier (KM) Survival analysis of MAN2A1 was performed by sangerbox tool. The clinicopathologic features of MAN2A1 were evaluated with chi-squared, univariate, and logistic regression method by SPSS. Patients’ overall survival (OS)-related clinical characteristics in TCGA was analyzed by Cox regression via SPSS.
Results
Identification of hub genes in CRC
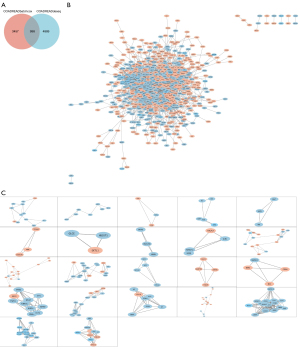
We drew a flow chart of research design (Figure 1). We downloaded expression profiles and clinical data of COAD and READ from TCGA and identified 4,455 genes associated with prognosis in CRC by cox analysis in batch mode (P≤0.05), identified 5,887 genes differentially expressed between metastatic CRC tissues and in situ tissues by edgR in Sangerbox tool (P≤0.05). Finally, we got 998 potential aim genes met the above requirements shown in the intersection of Venn diagram (Figure 2A). PPI network was constructed by STRING and cytoscape software for the 998 potential aim genes, composed of 551 up-regulated genes and 447 down-regulated genes (Figure 2B). Then we utilized MCODE plus to find 196 hub genes (central nodes), including 63 up-regulated genes and 133 down-regulated genes (Figure 2C, Table S1).
GO and signaling pathway enrichment analysis
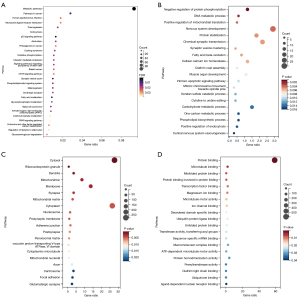
We analyzed the potential aim genes via the KOBAS online software and identified 28 significant enrichment pathways, indicating they were enriched in metabolic pathways, pathways in cancer, human papillomavirus interaction, endocytosis, P53 signaling pathway and so on (Figure 3A). And the DAVID online software was used to identify 113 significant enrichment GO terms, including biological process (BP), molecular function (MF), and cellular components (CC). The top 20 GO terms are depicted in Figure 3B-3D.
The expression of MAN2A1 in CRC
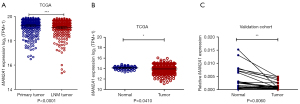
We found that the expression of MAN2A1 in CRC and metastatic CRC have not been reported. In addition, the relation between MAN2A1 expression and CRC survival was still controversial. Consequently, we explored the expression of MAN2A1 between metastatic CRC and non-metastatic CRC in TCGA database and found that MAN2A1 was lower expressed in metastatic CRC (Figure 4A). We also found the expression of MAN2A1 in TCGA and our own CRC tissues was down-expressed than adjacent normal tissues cohort (n=26, 16 of 26 were metastatic patients, 10 of 26 were nonmetastatic patients) (Figure 4B,4C).
Prognostic value of MAN2A1 expression in CRC
We evaluated the prognostic value of MAN2A1 in TCGA CRC dataset by performing KM survival analysis. The results showed that high expression of MAN2A1 was associated with better OS (Figure 5, P=0.0019). Univariate cox analysis was used to analyze independent prognostic value of MAN2A1, and the results showed that high expression of age (P=0.001), invasion depth (P=0.005), LNM (P<0.001), distant metastasis (P<0.001), and Tumor Node Metastasis (TNM) stage (P<0.001) corresponded with poor OS in CRC patients, whereas MAN2A1 (P=0.013), radiation treatment (P=0.047) corresponded with favorable OS in CRC patients (Table 2).
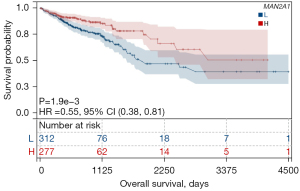
Table 2
| Parameter | Univariate analysis | ||
|---|---|---|---|
| P value | HR | 95% CI | |
| Age (≥65 vs. <65), years | 0.001** | 1.984 | 1.34–2.938 |
| Gender (female vs. male) | 0.847 | 0.966 | 0.682–1.370 |
| Invasion depth (T3 & T4 vs. T1 & T2) | 0.005 | 2.444 | 1.314–4.547 |
| Lymph node metastasis (yes vs. no) | <0.001*** | 2.509 | 1.749–3.599 |
| Distant metastasis (yes vs. no) | <0.001*** | 3.766 | 2.551–5.561 |
| TNM stage (stages III & IV vs. Stages I & II) | <0.001*** | 2.832 | 1.936–4.143 |
| MAN2A1 expression (high vs. low) | 0.013 | 0.637 | 0.446–0.909 |
| Pharmacological treatment (yes vs. no) | 0.731 | 1.089 | 0.670–1.770 |
| Radiation treatment (yes vs. no) | 0.047 | 0.592 | 0.353–0.992 |
**, P<0.01; ***, P<0.001. CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; HR, hazard ratio; TNM, Tumor Node Metastasis.
The association of MAN2A1 expression and clinicopathologic characteristics in CRC
The expression level of MAN2A1 was associated with the prognosis in CRC patients. The expression of MAN2A1 was significantly related to LNM (P=0.05), and TNM stage (P=0.02) (Table 3). Whereas age, gender, radiation treatment, pharmacological treatment, invasion depth, and distant metastasis were not correlate with MAN2A1 expression (Table 3). Logistic regression analysis results showed that the expression of MAN2A1 was associated with LNM [yes vs. no, odd ratio (OR): 0.725, 95% CI: 0.526–1.001, P=0.05], distant metastasis (yes vs. no, OR: 0.634, 95% CI: 0.396–1.015, P=0.058), and TNM stage (stages III and IV vs. stages I and II, OR: 0.684, 95% CI: 0.494–0.947, P=0.022) (Table 4).
Table 3
| Variables | Total | Low expression | High expression | χ2 | P |
|---|---|---|---|---|---|
| Age, years | 0.902 | 0.342 | |||
| <65 | 249 | 127 | 122 | ||
| ≥65 | 368 | 202 | 166 | ||
| Gender | 0.772 | 0.380 | |||
| Female | 288 | 159 | 129 | ||
| Male | 329 | 170 | 159 | ||
| Radiation treatment | 1.970 | 0.160 | |||
| Yes | 128 | 72 | 56 | ||
| No | 393 | 193 | 200 | ||
| Pharmacological treatment | 0.835 | 0.361 | |||
| Yes | 124 | 66 | 58 | ||
| No | 395 | 195 | 200 | ||
| TNM stage | 5.426 | 0.02* | |||
| Stage I | 105 | 55 | 50 | ||
| Stage II | 226 | 108 | 118 | ||
| Stage III | 179 | 101 | 78 | ||
| Stage IV | 88 | 56 | 32 | ||
| Invasion depth | 0.012 | 0.914 | |||
| T1 | 20 | 10 | 10 | ||
| T2 | 105 | 57 | 48 | ||
| T3 | 420 | 228 | 192 | ||
| T4 | 70 | 32 | 38 | ||
| Lymph node metastasis | 3.827 | 0.05* | |||
| Yes | 264 | 153 | 111 | ||
| No | 350 | 175 | 175 | ||
| Distant metastasis | 3.640 | 0.056 | |||
| Yes | 88 | 56 | 32 | ||
| No | 464 | 244 | 220 | ||
*, P<0.05. CRC, colorectal cancer; TCGA, The Cancer Genome Atlas.
Table 4
| Logistic regression analysis | P | HR | 95% CI |
|---|---|---|---|
| Lymph node metastasis (yes vs. no) | 0.05* | 0.725 | 0.526–1.001 |
| Distant metastasis (yes vs. no) | 0.058 | 0.634 | 0.396–1.015 |
| TNM stage (stages III & IV vs. stages I & II) | 0.022* | 0.684 | 0.494–0.947 |
*, P<0.05. CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; HR, hazard ratio; TNM, Tumor Node Metastasis.
KEGG and GO pathway enrichment analysis by GSEA
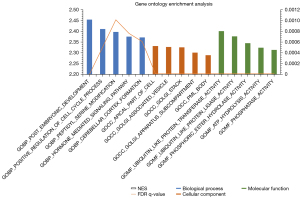
The top 20 signaling pathways enriched in the MAN2A1 high-expression group were showed in Table 5. Enriched top pathways such as WNT signaling pathway, ubiquitin mediated proteolysis, CRC, pathways in cancer, prostate cancer, TGF beta signaling pathway, MAPK signaling pathway, ERBB signaling pathway and apoptosis were associated with cancer occurrence (Table 5). Gene Ontology contains biological process, cellular components, and molecular function. The enriched top 5 for each category are shown in Figure 6. The enriched biological process were post embryonic development, positive regulation of cell cycle process, peptidyl serine modification, hormone mediated signaling pathway and cerebellar cortex formation. The enriched cellular components were apical part of cell, golgi associated vesicle, golgi stack, golgi apparatus subcompartment and pml body. The enriched molecular functions were ubiquitin like protein transferase activity, ubiquitin like protein ligase activity, phosphoric ester hydrolase activity, adenosine-triphosphate (ATP) hydrolysis activity and phosphatase activity.
Table 5
| Signaling pathway | NES | NOM p-value | FDR q-value |
|---|---|---|---|
| KEGG_wnt signaling pathway | 2.361 | ≤0.001 | 0.003 |
| KEGG_long term potentiation | 2.328 | ≤0.001 | 0.001 |
| KEGG_oocyte meiosis | 2.296 | ≤0.001 | 9.30E-04 |
| KEGG_ubiquitin mediated proteolysis | 2.281 | ≤0.001 | 6.98E-04 |
| KEGG_neurotrophin signaling pathway | 2.259 | ≤0.001 | 0.001 |
| KEGG_GNRH signaling pathway | 2.228 | ≤0.001 | 0.001 |
| KEGG_progesterone mediated oocyte maturation | 2.223 | ≤0.001 | 9.62E-04 |
| KEGG_colorectal cancer | 2.223 | ≤0.001 | 8.42E-04 |
| KEGG_pathways in cancer | 2.211 | ≤0.001 | 7.48E-04 |
| KEGG_prostate cancer | 2.201 | ≤0.001 | 6.73E-04 |
| KEGG_TGF beta signaling pathway | 2.195 | ≤0.001 | 6.12E-04 |
| KEGG_lysine degradation | 2.193 | ≤0.001 | 5.61E-04 |
| KEGG_MAPK signaling pathway | 2.184 | ≤0.001 | 5.18E-04 |
| KEGG_ERBB signaling pathway | 2.176 | ≤0.001 | 7.73E-04 |
| KEGG_apoptosis | 2.176 | ≤0.001 | 7.22E-04 |
| KEGG_melanogenesis | 2.174 | ≤0.001 | 7.41E-04 |
| KEGG_butanoate metabolism | 2.171 | ≤0.001 | 6.97E-04 |
| KEGG_phosphatidylinositol signaling system | 2.171 | ≤0.001 | 6.58E-04 |
| KEGG_P53 signaling pathway | 2.169 | ≤0.001 | 6.24E-04 |
| KEGG_beta alanine metabolism | 2.161 | ≤0.001 | 7.03E-04 |
NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate.
Discussion
As the World Health Organization (WHO) reported, the incidence and mortality rate of CRC ranks third in the United States (19). Previous studies have shown kinds of mechanisms of CRC, including chromosomal instability (20,21), microsatellite instability, oncogenes signaling pathways, metabolic alterations (22,23), abnormal immune response (24-26) etc. Numerous genes have been considered as prognostic predictors and potential therapeutic targets of CRC, the genes associated with CRC metastasis are in same significance.
The development of microarray technology and bioinformatics science provides an advanced method to solve clinical problems. In the present study, we screened TCGA-COAD and TCGA-READ datasets to identify genes significantly correlated with prognosis, and 4,455 genes were identified. The 4,455 genes were further analyzed to identify potential aim genes in both COAD and READ datasets, and we finally obtained 998 genes.
In addition, we constructed a PPI network with STRING online tools and analyzed with Cytoscape software. We also analyzed enrichment in GO and KEGG signaling pathways and found that they were enriched in 28 pathways, such as metabolic pathways, pathways in cancer, human papillomavirus interaction, endocytosis, P53 signaling pathway and so on which were relate to cancer occurrence and progress. Then, we used Cytoscape MCODE plus to analyze the 998 genes to get functional modules and identify hub genes based on topology to confirm densely connected regions. And find 196 hub genes (central nodes) that included 63 up-regulated genes and 133 down-regulated genes.
The metabolic reprogramming is related to the initiation, progression and metastasis of carcinomas, including CRC. Cancer cells usually need more nutrients, redox and energy to support their rapid proliferation and division. Cancer cells supply what they need through abnormal glycolysis, glutamine, glutaminolysis, serine and tryptophan metabolism, one-carbon metabolism and lipid metabolism (27). Previous studies have shown that the metabolic reprogramming is regulated by expression of oncogenic pathways and tumor suppressor genes, such as WNT signaling, KRAS signaling, PI3K/AKT/mTOR signaling, P53 signaling and so on (22,28).
In this study, we detected the expression of DEGs at the mRNA level afterwards and found MAN2A1 had lower expression levels in CRC than adjacent normal tissue; in addition, the expression of MAN2A1 was down-expressed in CRC tissues than normal tissues and down-expressed in metastatic CRC than non-metastatic CRC in TCGA. And the increasing expression of MAN2A1 was significantly negatively associated with CRC progression. Hence, we regarded MAN2A1 as a CRC-relevant gene.
MAN2A1 is a Golgi enzyme converting high mannose to complex type N-glycan for maturing membrane protein glycosylation (29,30). MAN2A1 fuses FER and turns to oncogene, previous analyses have shown that about 80% prostate cancer patients with MAN2A1-FER is positive associated with poor clinical outcome (31,32), other analysis has found that MAN2A1-FER fusion occur in liver cancer, esophageal cancer and other types malignancies (33). MAN2A1-FER fusion translocates FER kinase to Golgi apparatus, FER kinase activates and promote cancers through epidermal growth factor receptor (EGFR) signaling pathway (34-36).
In this study, via data mining in TCGA and validating in our cohort, we found MAN2A1 was significantly downregulated in CRC tissues than adjacent normal tissues and downregulated in metastatic CRC than non-metastatic CRC. These findings suggested that MAN2A1 might be inhibited in CRC cases. Furthermore, high expression of MAN2A1 was negatively and significantly related to TNM stage and LNM. The Kaplan-Meier survival analysis and COX analysis indicated that high expressed MAN2A1 was associated with better OS. Furthermore, GSEA analysis found MAN2A1 expression was related to wnt signaling pathway, CRC pathway, pathways in cancer, TGF beta signaling pathway, MAPK signaling pathway, P53 signaling pathway and so on. These results indicated that potential biological mechanism of MAN2A1 may be relate to CRC occurrence and progress. Nevertheless, the mechanisms of MAN2A1 in CRC need further study to explain. These results would be helpful for providing candidate targets for the diagnosis and prognosis for CRC patients, as well as new treatment strategy.
Acknowledgments
We sincerely thank the staff of Sir Run Run Shaw Hospital, affiliated to Zhejiang University for their laboratory and technical support.
Funding: The study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-629/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-629/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-629/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Sir Run Run Shaw Hospital, affiliated to Zhejiang University approved all study protocols (approval ID. 20200619-34). Individual consent for this retrospective study was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252-71. [Crossref] [PubMed]
- Field K, Lipton L. Metastatic colorectal cancer-past, progress and future. World J Gastroenterol 2007;13:3806-15. [Crossref] [PubMed]
- Shih W, Chetty R, Tsao MS. Expression profiling by microarrays in colorectal cancer Oncol Rep 2005;13:517-24. (Review). [Crossref] [PubMed]
- Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol 2006;24:971-83. [Crossref] [PubMed]
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Wu J, Mao X, Cai T, et al. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res 2006;34:W720-4. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Schmid F, Schmid M, Müssel C, et al. GiANT: gene set uncertainty in enrichment analysis. Bioinformatics 2016;32:1891-4. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739-40. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, et al. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci 2017;18:197. [Crossref] [PubMed]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010;7:153-62. [Crossref] [PubMed]
- La Vecchia S, Sebastián C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin Cell Dev Biol 2020;98:63-70. [Crossref] [PubMed]
- Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015;149:1204-1225.e12. [Crossref] [PubMed]
- Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361-75. [Crossref] [PubMed]
- Johdi NA, Sukor NF. Colorectal Cancer Immunotherapy: Options and Strategies. Front Immunol 2020;11:1624. [Crossref] [PubMed]
- Lichtenstern CR, Ngu RK, Shalapour S, et al. Immunotherapy, Inflammation and Colorectal Cancer. Cells 2020;9:618. [Crossref] [PubMed]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23:27-47. [Crossref] [PubMed]
- Nenkov M, Ma Y, Gaßler N, et al. Metabolic Reprogramming of Colorectal Cancer Cells and the Microenvironment: Implication for Therapy. Int J Mol Sci 2021;22:6262. [Crossref] [PubMed]
- Shi S, Gu S, Han T, et al. Inhibition of MAN2A1 Enhances the Immune Response to Anti-PD-L1 in Human Tumors. Clin Cancer Res 2020;26:5990-6002. [Crossref] [PubMed]
- Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13:397-406. [Crossref] [PubMed]
- Yu YP, Liu S, Nelson J, et al. Detection of fusion gene transcripts in the blood samples of prostate cancer patients. Sci Rep 2021;11:16995. [Crossref] [PubMed]
- Yu YP, Ding Y, Chen Z, et al. Novel fusion transcripts associate with progressive prostate cancer. Am J Pathol 2014;184:2840-9. [Crossref] [PubMed]
- Chen ZH, Yu YP, Tao J, et al. MAN2A1-FER Fusion Gene Is Expressed by Human Liver and Other Tumor Types and Has Oncogenic Activity in Mice. Gastroenterology 2017;153:1120-1132.e15. [Crossref] [PubMed]
- Lee CC, Shiao HY, Wang WC, et al. Small-molecule EGFR tyrosine kinase inhibitors for the treatment of cancer. Expert Opin Investig Drugs 2014;23:1333-48. [Crossref] [PubMed]
- Gala K, Chandarlapaty S. Molecular pathways: HER3 targeted therapy. Clin Cancer Res 2014;20:1410-6. [Crossref] [PubMed]
- Luo SY, Lam DC. Oncogenic driver mutations in lung cancer. Transl Respir Med 2013;1:6. [Crossref] [PubMed]


