
LPCAT3 is a potential prognostic biomarker and may be correlated with immune infiltration and ferroptosis in acute myeloid leukemia: a pan-cancer analysis
IntroductionOther Section
LPCAT3 is known as membrane-bound O-acyltransferase 5 (MBOAT5), which was initially shown to play a key role in the reacylation step of the catalytic phospholipid remodeling process, also known as the Lands cycle (1-4). It is vital for biological function in lipid metabolism (5,6). A growing body of evidence suggests that LPCAT3 plays a central role in non-apoptotic cell death, especially ferroptosis (7,8). Ferroptosis is characterized by intracellular iron overload and lipid peroxides (9). Polyunsaturated fatty acid-containing phospholipids are the main substrates of lipid peroxidation in ferroptosis, which is positively regulated by enzymes, such as acyl-CoA synthetase long-chain family member 4 (ACSL4), LPCAT3, lipoxygenases (ALOXs), or cytochrome P450 oxidoreductase (POR). In particular, ACSL4 and LPCAT3 play a key role in promoting ferroptosis by incorporating polyunsaturated fatty acids (PUFAs) into cellular phospholipids (especially phosphatidylethanolamine) (10-12). Therefore, LPCAT3 is considered to be one of the driver genes that promote ferroptosis. In addition, LPCAT3 may pro-tumorigenic activity of several neoplasms (13), including ovarian cancer (OV) (14) and acute myeloid leukemia (AML) (15), which means that LPCAT3 has potential clinical translational value in cancer patients. Therefore, it is particularly important to study the regulatory function and molecular mechanism of LPCAT3 in pan-cancer datasets to provide new directions and strategies for the clinical treatment of cancer.
However, little literatures have reported the expression level of LPCAT3 in different classes of cancer and its impact on the clinical significance of cancer in terms of biological function. This present study used The Cancer Genome Atlas (TCGA) database to explore the expression profile and prognostic value of LPCAT3 in 33 types of cancer. Concomitantly, we reported the results of LPCAT3 analysis in AML. We observed that LPCAT3 widely expressed across many cancers and affects the prognosis of patients by affecting infiltrating immune cells and ferroptosis. This investigation offers a fresh perspective on how LPCAT3 affects prognosis in pan-cancer by modulating the tumor immune environment through lipid metabolism leading to ferroptosis of tumor-suppressing associated immune cells. We present the following article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-985/rc).
MethodsOther Section
Description of the analysis tools
The TIMER2.0 web server provides comprehensive analysis and visualization functions of tumor infiltrating immune cells (http://timer.cistrome.org/) (16). The OPEN TARGET platform (https://www.targetvalidation.org/) integrates genetics, omics, and chemical data to identify the involvement of genes in diseases and aid systematic drug target identification and prioritization (17). Enrichr (https://maayanlab.cloud/Enrichr/enrich#) is a web server for several enrichment analyses of gene sets (18,19). The data of 33 cancer types were collected from the GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/), an online tool, including genomic and immunogenomic data, drug responses data, and normal tissue data (20). The ROCPlotter (http://www.rocplot.org/) is a transcriptome-based tool for predicting biomarkers by linking gene expressions and responses to therapy of breast, ovarian, colorectal, and glioblastoma cancer patients (21). We used the STRING database (https://cn.string-db.org/) to construct the protein-protein interaction network and investigate interacting genes (22). The GeneMANIA (http://genemania.org/) is a web server as biological network integration for gene prioritization and predicting gene function (23). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data collection
The clinical data and gene expression profiles data of cancer cohorts were downloaded from the TCGA database. The detailed information of the 33 TCGA cancers is shown in Table S1. However, the gene expression profiles of the healthy individuals were downloaded from The Genotype-Tissue Expression (GTEx) (https://www.gtexportal.org/home/). All the data used in this study complied with the publication guidelines stipulated by TCGA and GTEx database, as a result, ethical approval and informed consent are not required.
LPCAT3 expression pattern in pan-cancer
Based on the TCGA and GTEx database, we analyzed the differential expression of LPCAT3 between various types of cancer and normal tissues. All expression data were normalized via log2 transformation.
Prognostic analysis
According to the median mRNA expression level of LPCAT3, patients were divided into high- and low-LPCAT3 expression groups. We compared the prognosis between the different LPCAT3 expression groups and the 33 types of cancer, including overall survival (OS), disease-specific survival (DSS), and progression-free survival (PFS). The difference in survival between groups was tested by the log-rank test (P<0.05). Prognostic genes were then screened using univariate Cox regression analysis (P<0.05).
Relationship between LPCAT3 expression and tumor immune microenvironment (TIME) analysis
Immune score, microenvironment score, and stromal score analysis were performed by the “immunedeconv” R package, which included TIMER, XCell, MCP-counter, CIBERSORT, EPIC, and quantized. The detailed algorithm of the immune score was reported according to the previously described (24,25). We apply the TIMER algorithm to estimate 6 types of infiltrating immune cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, and then evaluated the relationship between the LPCAT3 expression and the levels of 6 types of infiltrating immune cells. To ensure the accuracy of our results, we also utilized the XCell algorithm to estimate 35 types of infiltrating immune cells, then, we compared the levels of 35 types of infiltrating immune cells with the LPCAT3 expression.
Correlations of LPCAT3 expression with check-point molecules and immunological associated genes
We also explored the relationship between the expression level of 8 immune checkpoints (SIGLEC15, IDO1, CD274, HAVCR2, PDCD1, CTLA4, LAG3, and PDCD1LG2) and the level of LPCAT3 expression. Furthermore, the correlation analysis was also performed between the expression level of LPCAT3 and the level of immune checkpoint-related genes.
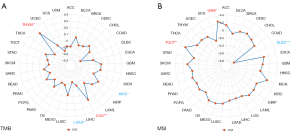
Relationship between the LPCAT3 expression and tumor mutational burden (TMB) as well as microsatellite instability (MSI)
TMB can reflect the number of mutations in tumor cells, so it has been used as a quantifiable immune-response biomarker. We used the somatic mutation data, downloaded from TCGA, to calculate the TMB scores. MSI scores were obtained for all samples based on somatic mutation data downloaded from TCGA. The correlation analysis between LPCAT3 expression and TMB as well as MSI was performed by Spearman’s method (26). In our study, the red plot in the figure represents the correlation coefficient between LPCAT3 and TMB or MSI.
Functional enrichment analysis
We used Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analysis (GSEA) analysis to investigate the biological, molecular function, and potential molecular mechanism of LPCAT3 in cancer. A total of 128 genes were used for GO and KEGG enrichment analysis, which were downloaded from the GEPIA2 website (100 similar genes) and STRING website (28 adjacent genes). To explore the potential biological functions of the LPCAT3 between high- and low-LPCAT3 expression in AML, we conducted GSEA based on the curated gene sets c2 kegg symbols. GO, KEGG, and GSEA analysis was performed using the “ClusteProfiler” package in R (27).
Estimation of chemotherapy drug response
Based on the median cut-off of LPCAT3 expression, patients were divided into high- and low-LPCAT3 expression groups. The half-maximal inhibitory concentration (IC50) values of chemotherapy drugs (BCL2 inhibitors, Midostaurin and Sorafenib), from the Genomics of Drug Sensitivity in Cancer (GDSC) project (www.cancerRxgene.org), were predicted by the “pRRophetic” package, and then was compared the drug response between the different expression groups.
Statistical analysis
All analyses were conducted using the statistical software R (version 4.1). Alterations in LPCAT3 expression levels in cancer tissues and normal tissues were analyzed by Wilcoxon rank-sum test. The median expression level of LPCAT3 was regarded as the cut-off value. The prognostic factors were evaluated by Cox regression analysis and the Kaplan-Meier method. The effect of risk factors on OS was evaluated by the Cox proportional hazards regression model. Candidate variables with P<0.05 in univariate analysis were reserved and incorporated in multivariate analysis. The P value was set at the routine 0.05 significance level.
ResultsOther Section
LPCAT3 gene structure, single-cell localization, variations, and expression profiles under physiological conditions
LPCAT3, also known as MBOAT5, is located on chromosome 12p13.31, containing 13 exons (Figure 1A). The distribution of LPCAT3 in the endoplasmic reticulum (ER) and microtubules using GeneCards database (Figure 1B). The LPCAT3 protein topology revealed a crossing membrane (Figure 1C). Furthermore, we observed LPCAT3 messenger mRNA expression in various normal immune cells and tissues (Figure 1D,1E). Interestingly, LPCAT3 is highly expressed in monocytes. LPCAT3 is associated with a variety of cancers, metabolic diseases, immune system, and hematological diseases through gene-disease network (https://platform.opentargets.org/) interaction analysis (Figure 1F).
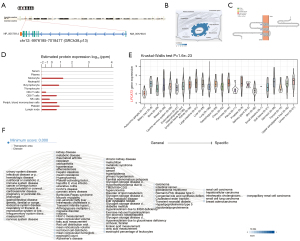
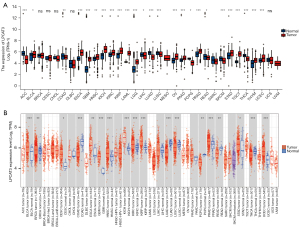
Pan-cancer expression landscape of LPCAT3
According to the data, downloaded from the TCGA database and GTEx, to compare the LPCAT3 expression. The expression level of LPCAT3 was significantly higher in tumor tissues versus adjacent tissues in the bladder urothelial carcinoma (BLCA), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), AML, lower-grade glioma (LGG), OV, pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thymoma (THYM), and uterine corpus endometrial carcinoma (UCEC) (Figure 2A). However, the expression level of LPCAT3 was observed to increase in tumor tissues versus normal tissues in the BLCA, breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), ESCA, GBM, HNSC, KICH, KIRC, KIRP, PRAD, STAD, and UCEC based on the TIMER database (Figure 2B).
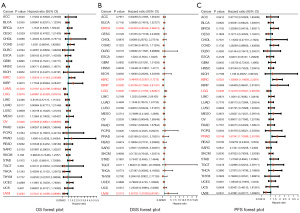
Prognostic value of LPCAT3 in pan-cancer
In pan-cancer data, we assessed the relationship between LPCAT3 expression and prognosis, including OS, DSS, and PFS. Univariate Cox analysis to conclude that LPCAT3 expression was significantly associated with OS in 5 cancer types, including KIRC, AML, LGG, OV, and uveal melanoma (UVM) (Figure 3A). The Kaplan-Meier survival curve showed that the upregulation of LPCAT3 expression was significantly correlated with poorer OS in AML, LGG, OV, and UVM, while the opposite in KIRC. (Figure S1). Moreover, the relationship between LPCAT3 expression and DSS in cancer patients was investigated. The results of COX regression analysis showed that LPCAT3 expression was associated with DSS in BRCA, KIRC, KIRP, LGG, and UVM (Figure 3B). Kaplan-Meier analysis revealed that increased expression of LPCAT3 correlated with poor DSS in LGG and UVM patients, while increased LPCAT3 expression predicted good DSS in BRCA, KIRC, and KIRP (Figure S2). We further analyzed the association between the LPCAT3 expression and PFS in pan-cancer. The results of COX regression showed that the expression level of LPCAT3 was significantly correlated with LGG, KIRC, and PRAD (Figure 3C). The Kaplan-Meier analysis showed that compared with the low-LPCAT3 expression patients, the high-LPCAT3 expression patients were correlated with a worse PFS in LGG, while a better PFS in KIRC and PRAD (Figure S3). In addition, adjusted by other covariates, multivariate analysis confirmed that high-LPCAT3 expression adversely impacted OS (HR, 2.183, 95% CI: 1.34–3.57; P=0.002) in AML (Table S2).
Relationship between LPCAT3 expression and the tumor immune microenvironment
The previous study has indicated that the quantity and activity status of tumor immune cells are the important predictive criterion for cancer survival times (28). Hence, we evaluated the correlation between immune infiltration level and LPCAT3 expression. First, based on the TIMER algorithm, we analyzed the relationship between LPCAT3 expression and the levels of 6 types of infiltrating immune cells. Figure 4A showed that LPCAT3 expression was significantly correlated with immune infiltrating cell abundance. LPCAT3 level was significantly correlated with the infiltration levels of CD8+ T cells in 15 cancer types (such as THCA, PAAD, PRAD, etc.), CD4+ T cells in 14 cancer types (such as COAD, HNSC, READ, etc.), neutrophils in 19 cancer types (such as DLBC, LGG, PRAD, etc.), DCs in 18 cancer types (such as COAD, LGG, PRAD, etc.), macrophages in 20 cancer types (such as THCA, THYM, PRAD, etc.), and B cells in 11 types of cancer (such as KIRC, PCPG, READ, etc.) (all P<0.05). Besides, in our study, to ensure the accuracy of our results, we also utilized the XCell algorithm to estimate 35 types of infiltrating immune cells and 3 kinds of TIME scores. we observed that NK/T cell, CD4+ T cell (Th1 cell), and Plasmacytoid dendritic cells were negatively associated with the LPCAT3 expression in pan-cancer, while Mast cell and Common lymphoid progenitor were positively correlated with the LPCAT3 expression in Pan-cancer (Figure 4B). Moreover, T cell CD4+ effector memory, Eosinophils, Common myeloid progenitor cell, and B cell plasma were negatively correlated with the LPCAT3 expression in AML. However, Macrophages M2, followed by Monocytes, Macrophages M1, Macrophages, and myeloid dendritic activated cells were positively associated with the LPCAT3 expression in AML. (Figure 4B). In addition, our results showed that LPCAT3 expression positively correlated with immune score and microenvironment score in AML and LGG, while negatively correlated with an immune score, microenvironment score, and stromal score in multiple cancers, including BLCA, BRCA, KICH, KIRC, KIRP, LUSC, PCPG, PRAD, TGCT, THCA, THYM, and UCEC (Figure 4B). The result of correlation analysis indicated that the monocytes (r=0.32), monocyte macrophages (r=0.32), and neutrophils (r=0.30) were positively correlated with the LPCAT3 expression, while the T cell was negatively correlated with the LPCAT3 expression (r=−0.20) (Figure 4C).
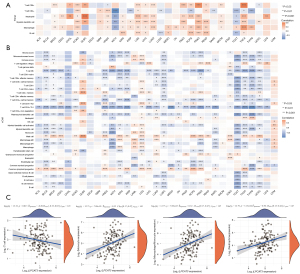
Correlations of LPCAT3 expression levels with check-point molecules and immunological associated genes
To estimate the relationship between the LPCAT3 expression and the potential therapeutic value of the immune checkpoint, we evaluated the association between the LPCAT3 expression and eight immune checkpoints (TIGIT, SIGLEC15, PDCD1LG2, PDCD1, LAG3, HAVCR2, CTLA4, and CD274). Notably, we observed that the expression level of LPCAT3 was negatively correlated with most immune checkpoints in ACC, LUSC, TGCT, and THCA. In contrast, the expression level of LPCAT3 was positively correlated with most checkpoints in COAD, DLBC, LGG, LIHC, and UVM (Figure 5A). In addition, the results indicated that only two immune checkpoints of PDCD1LG2 and CD274 were positively correlated with the expression of LPCAT3 (Figure 5A). We further investigated the correlation between the LPCAT3 expression and immunological associated genes through the GEPIA2 tool and found that it was positively related to CD27, CD40, CD80, CD274, CTLA4, PDCD1LG2, TIGIT, and etc. In contrast, a negatively correlation was found between LPCAT3 expression and IMIGD2 as well as CD244 (Figure 5B).

The relationship between the LPCAT3 expression and TMB as well as MSI score
Previous research indicated that TMB and MSI are considered essential factors impacting the occurrence and progression of the tumor (29). So, it is necessary to further explore the relationship between LPCAT3 expression and TMB as well as MSI score. The results demonstrated that the LPCAT3 expression was positively associated with the TMB score in LGG and THYM, while, the LPCAT3 expression was negatively associated with the TMB score in KIRC and LUAD (Figure 6A). Furthermore, the LPCAT3 expression was positively correlated with MSI in UVM and TGCT, and negatively correlated with MSI in DLBC (Figure 6B).
LPCAT3-related gene enrichment analysis
To analyze the molecular mechanism of the LPCAT3 gene in tumorigenesis, we screened out the targeting LPCAT3-binding proteins and the LPCAT3 expression-related genes for pathway enrichment. Figure 7A,7B showed that the interaction network of LPCAT3 has 25 binding proteins based on the STRING tool and GeneMania. We also used the GEPIA2 tool to obtain the top 100 genes that are remarkably similar to LPCAT3 expression. Among the 100 similar genes, we selected five genes (BRAP, HK1, TLN1, TPD52L2, and PNPO) that were most associated with LPCAT3. As shown in Figure 7C-7G, the expression level of LPCAT3 was significantly correlated with that of BRAP (R=0.73), HK1 (R=0.65), TLN1 (R=0.6), TPD52L2 (R=0.6), and PNPO (R=0.62) genes (all P<0.001).
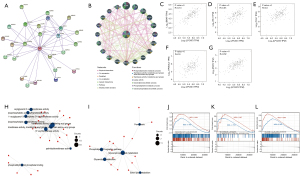
Furthermore, we also downloaded 28 adjacent genes of LPCAT3 from the STRING. To better understand the functional implication of the 128 genes (100 similar genes and 28 adjacent genes), the GO terms of BP, CC, and MF, as well as KEGG for those genes were explored. The results showed that GO enrichment analysis was primarily enriched in the pathways of lipid metabolism or cellular biology, such as O-acyltransferase activity lysophospholipid acyltransferase activity, transferring acyl groups, lysophosphatidic acid acyltransferase activity, transferase activity, and others (Figure 7H). KEGG pathway analysis revealed that 128 genes were mainly enriched in the phospholipase D signaling pathway, Lipid metabolism, and Ferroptosis processes (all adjusted P<0.05) (Figure 7I). The KEGG enrichment analysis indicated that the lipid metabolism and ferroptosis processes might be involved in the effect of LPCAT3 on tumorigenesis.
To investigate the differential activation of LPCAT3-related signaling pathways in cancer, the GSEA analysis was performed. GSEA analysis in the KEGG gene set demonstrated that the genes are enriched in pathways like the FLT3 signaling pathway, PD-1 signaling pathway, IL6-JAK-STAT3 signaling pathway, hematopoietic stem cell differentiation, fatty acid metabolism, and adipogenesis (Figure 7J-7L).
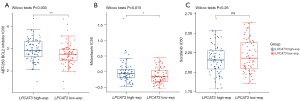
Drug response between high- and low-LPCAT3 expression groups
We estimated the drug sensitivity between the different LPCAT3 expression groups with BCL2 inhibitors, Midostaurin, and Sorafenib in the TCGA AML cohort. In comparison to the low-LPCAT3 expression group, there was a prominently increased IC50 value of BCL2 inhibitors (P=0.003) and midostaurin (P=0.015) in high-LPCAT3 expression group (Figure 8A,8B). This indicates that low-risk patients displayed more benefit to BCL2 inhibitors and Midostaurin. However, the sensitivity of sorafenib was not significantly different between the high- and low-LPCAT3 expression groups (Figure 8C).
DiscussionOther Section
In recent years, with the improvement of transplantation technology and supportive therapy, the overall mortality rate of AML has decreased slightly, but the survival rate is still not satisfactory (30). Early detection and effective treatment measures are essential for enhancing the prognosis of AML. To identify early diagnosis and sensitive biomarkers of cancers, more and more pan-cancer analysis studies shape genetic mutations and cancer driver genes (31,32). This study reveals the similarities and differences of LPCAT3 among different cancers through pan-cancer analysis and provides potential personalized treatment strategies for AML.
LPCAT3 is involved in the reacylation process of the lysophosphatidic transferase (LPLATs) catalyzed phospholipid remodeling pathway in the liver, namely, the Lands cycle (2). LPCAT3 is known to be overexpressed in nasal epithelium and liver and up-regulated in Monocytes, B lymphocytes, CD8+ T cells, NK cells, CD4+ T cells, and Lymph nodes (33). However, its roles in pan-cancer and whether it can be used as a biomarker are still unclear.
In this study, we utilized GTEx and TCGA databases to detect expression level of LPCAT3 and its effects on prognosis in human pan-cancer. We found for the first time that LPCAT3 is abnormally overexpressed in multiple cancers including BLCA, BRCA, ESCA, CESC, GBM, HNSC, KICH, KIRC, KIRP, AML, LGG, OV, PAAD, PRAD, STAD, TGCT, THYM, and UCEC compared with adjacent tissues. This result is consistent with the previous study that LPCAT3 is overexpressed in BRCA tissues compared with their normal tissues (34). In addition, we also analyzed the relationship between LPCAT3 levels and the prognosis of patients in human pan-cancer. Our results showed that the up-regulation of LPCAT3 expression is associated with poor prognosis in several tumor types, including AML, LGG, OV, and UVM. However, up-regulation of LPCAT3 expression correlated with better prognosis in KIRC. The previous study also indicated the same result that an increased LPCAT3 expression correlated with poor prognosis in OV (35).
Evidence has accrued that a comprehensive understanding of the status of TIME in cancer patients is particularly important for selecting clinical-decision. To further improve our understanding of the mechanisms by which LPCAT3 expression may mediate differential prognosis in human pan-cancer, TIME showed that LPCAT3 significantly correlated with the immune infiltration levels of CD8+ T cells, CD4+ T cells, neutrophils, myeloid dendritic cell, macrophages, and B cell. We further used the XCell algorithm to examine the association between LPCAT3 expression and the immune infiltration levels of different types of immune cells. Our results showed that NK/T cell, CD4+ T cell (Th1 cell), and Plasmacytoid dendritic cells were negatively associated with the LPCAT3 expression in pan-cancer, while Mast cell and Common lymphoid progenitor were positively correlated with the LPCAT3 expression in pan-cancer. Furthermore, in the AML cohort, we found that the up-regulation of LPCAT3 expression was positively associated with the score of Macrophages M2. Macrophages are part of anti-tumor immunity. However, Macrophages M2 do not kill tumor cells but promote tumor development (36), which may help to explain to some extent why high-LPCAT3 expression AML patients with poor prognosis. Interestingly, our results showed that LPCAT3 expression was positively correlated with immune score and microenvironment score in AML and LGG, while negatively correlated with an immune score, microenvironment score, and stromal score in most cancer types. This suggests that a higher immune score may be associated with a poor prognosis in AML, which is consistent with the previous research (37).
The correlation between the LPCAT3 expression and immune checkpoints was also analyzed. We observed that LPCAT3 expression was positively correlated with immune checkpoints in various types of cancer, including COAD, DLBC, LGG, LIHC, OV, THYM, and UVM. These results strongly indicate that LPCAT3 may be a potential biomarker and play vital roles in tumor immunity. However, in the AML cohort, only two immune checkpoints of PDCD1LG2 and CD274 were positively correlated with the expression level of LPCAT3, those might help to explain why AML patients limited benefit from immune checkpoint therapy (38). In addition, we also found that the LPCAT3 expression was a significant correlation with immunological associated genes. These findings suggest that LPCAT3 plays an important role in regulating immunity in human pan-cancer.
TMB is an emerging pan-cancer predictive biomarker and can guide immunotherapy, which has been demonstrated in the non-small-cell lung (39) and colorectal (40) cancers. Furthermore, TMB can also predict prognosis after immunotherapy in human pan-cancer (41). MSI is also a promising biomarker for predicting immunotherapy response. Our study demonstrated that LPCAT3 expression is correlated with TMB in 4 cancer types (including THYM, LGG, KIRC, and LUAD) and with MSI in 3 cancer types (including TGCT, UVM, and DLBC). Consistent with previous research (42), our result also showed that the LPCAT3 expression will affect the TMB and MSI of multiple cancers, thereby affecting the patient's response to immune checkpoint therapy. However, further researches are needed to determine whether LPCAT3 can serve as a biomarker for predicting immunotherapy response in those cancers.
To further explore the molecular biological mechanism of the LPCAT3 in human pan-cancer, functional enrichment analysis showed that phospholipase D signaling pathway, Ferroptosis, and Lipid metabolism processes were found to be enriched. Ferroptosis is a programmed cell death process, which is marked by the accumulation of iron-dependent lipid peroxides. Recent studies have shown that reduced or increased sensitivity of ferroptosis can significantly promote AML tumor cell apoptosis (43,44). Ferroptosis promotes the production of immunosuppressive media by cancer cells and tumor-infiltrating immune cells, which may inhibit anti-tumor immunity and promote tumor growth (45). Furthermore, ferroptosis can also help adjacent cancer cells survive or evade immunity (46). Based on existing research and our findings, we infer that ferroptosis and immune microenvironment are significantly correlated, which together affect prognosis in pan-cancer, especially in AML. In addition, our study also showed that LPCAT3 expression was significantly associated with the lipid metabolism process, which was consistent with other LPCAT3 studies (1-3). In cancer, lipid metabolism is one of the most prominent metabolic changes, which can re-activate fat formation without relying on external lipids (46,47). These might be one of the reasons for the unfavorable prognosis in patients with overexpression of LPCAT3. In the AML cohort, the GSEA pathway analysis revealed that the FLT3 signaling pathway, PD-1 signaling pathway, IL6-JAK-STAT3 signaling pathway, hematopoietic stem cell differentiation, fatty acid metabolism, and adipogenesis were significantly activated, high-LPCAT3 expression group. The activation of the FLT3 signaling pathway has been reported as survival mechanism for drug resistance in AML (48). This might be one of the reasons for the poor prognosis in high-LPCAT3 expression group.
To evaluate patients may benefit from which drugs, we estimated the sensitivity of some chemotherapy drugs in AML. Our result indicated that the low-LPCAT3 expression patients were more likely to benefit from BCL2 inhibitor and midostaurin. This provides some references for our clinical drug selection and clinical trials.
In this research, we comprehensively analyzed the relationship between LPCAT3 expression and human pan-cancer. Nevertheless, there are several limitations in the current study. First, all the data came from public databases. Furthermore, the results of the study only came from bioinformatics analysis, further experimental are required to validate it and reveal the probable mechanism.
ConclusionsOther Section
The current study has indicated that LPCAT3 overexpression correlates with poor prognosis in multiple human cancers, including AML. Furthermore, we also tried to reveal the possible mechanism of the poor prognosis from many aspects. Based on the results of the present study, the LPCAT3 level is related to cancer immunity, ferroptosis, and lipid metabolism. These findings may provide a theoretical basis for the treatment AML via targeting ferroptosis and lipid metabolism. Therefore, LPCAT3 may serve as a potential prognostic and treatment biomarker.
AcknowledgmentsOther Section
We acknowledge the contributions of the TCGA and GTEx researchers.
Funding: This study was supported by grants from
FootnoteOther Section
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-985/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-985/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-985/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Gijón MA, Riekhof WR, Zarini S, et al. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem 2008;283:30235-45. [Crossref] [PubMed]
- Matsuda S, Inoue T, Lee HC, et al. Member of the membrane-bound O-acyltransferase (MBOAT) family encodes a lysophospholipid acyltransferase with broad substrate specificity. Genes Cells 2008;13:879-88. [Crossref] [PubMed]
- Zhao Y, Chen YQ, Bonacci TM, et al. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J Biol Chem 2008;283:8258-65. [Crossref] [PubMed]
- Jain S, Zhang X, Khandelwal PJ, et al. Characterization of human lysophospholipid acyltransferase 3. J Lipid Res 2009;50:1563-70. [Crossref] [PubMed]
- Schmidt JA, Brown WJ. Lysophosphatidic acid acyltransferase 3 regulates Golgi complex structure and function. J Cell Biol 2009;186:211-8. [Crossref] [PubMed]
- Li Z, Ding T, Pan X, et al. Lysophosphatidylcholine acyltransferase 3 knockdown-mediated liver lysophosphatidylcholine accumulation promotes very low density lipoprotein production by enhancing microsomal triglyceride transfer protein expression. J Biol Chem 2012;287:20122-31. [Crossref] [PubMed]
- Dixon SJ, Winter GE, Musavi LS, et al. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem Biol 2015;10:1604-9. [Crossref] [PubMed]
- Lee JY, Kim WK, Bae KH, et al. Lipid Metabolism and Ferroptosis. Biology (Basel) 2021;10:184. [Crossref] [PubMed]
- Sha W, Hu F, Xi Y, et al. Mechanism of Ferroptosis and Its Role in Type 2 Diabetes Mellitus. J Diabetes Res 2021;2021:9999612. [Crossref] [PubMed]
- Yuan H, Li X, Zhang X, et al. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun 2016;478:1338-43. [Crossref] [PubMed]
- Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 2017;13:91-8. [Crossref] [PubMed]
- Kagan VE, Mao G, Qu F, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol 2017;13:81-90. [Crossref] [PubMed]
- Di Conza G, Tsai CH, Gallart-Ayala H, et al. Tumor-induced reshuffling of lipid composition on the endoplasmic reticulum membrane sustains macrophage survival and pro-tumorigenic activity. Nat Immunol 2021;22:1403-15. [Crossref] [PubMed]
- Yang L, Tian S, Chen Y, et al. Ferroptosis-Related Gene Model to Predict Overall Survival of Ovarian Carcinoma. J Oncol 2021;2021:6687391. [Crossref] [PubMed]
- Song Y, Tian S, Zhang P, et al. Construction and Validation of a Novel Ferroptosis-Related Prognostic Model for Acute Myeloid Leukemia. Front Genet 2022;12:708699. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Carvalho-Silva D, Pierleoni A, Pignatelli M, et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res 2019;47:D1056-65. [Crossref] [PubMed]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [Crossref] [PubMed]
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90-7. [Crossref] [PubMed]
- Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 2018;34:3771-2. [Crossref] [PubMed]
- Fekete JT, Győrffy B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int J Cancer 2019;145:3140-51. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [Crossref] [PubMed]
- Wang J, Sun J, Liu LN, et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med 2019;25:656-66. [Crossref] [PubMed]
- Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol 2017; [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Dai S, Zeng H, Liu Z, et al. Intratumoral CXCL13+CD8+T cell infiltration determines poor clinical outcomes and immunoevasive contexture in patients with clear cell renal cell carcinoma. J Immunother Cancer 2021;9:e001823. [Crossref] [PubMed]
- Chida K, Kawazoe A, Kawazu M, et al. A Low Tumor Mutational Burden and PTEN Mutations Are Predictors of a Negative Response to PD-1 Blockade in MSI-H/dMMR Gastrointestinal Tumors. Clin Cancer Res 2021;27:3714-24. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Ju Q, Li X, Zhang H, et al. NFE2L2 Is a Potential Prognostic Biomarker and Is Correlated with Immune Infiltration in Brain Lower Grade Glioma: A Pan-Cancer Analysis. Oxid Med Cell Longev 2020;2020:3580719. [Crossref] [PubMed]
- Miao Y, Wang J, Li Q, et al. Prognostic value and immunological role of PDCD1 gene in pan-cancer. Int Immunopharmacol 2020;89:107080. [Crossref] [PubMed]
- Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017;547:453-7. [Crossref] [PubMed]
- Zhu L, Chen M, Huang B, et al. Genomic Analysis Uncovers Immune Microenvironment Characteristics and Drug Sensitivity of Ferroptosis in Breast Cancer Brain Metastasis. Front Genet 2022;12:819632. [Crossref] [PubMed]
- Wang H, Cheng Q, Chang K, et al. Integrated Analysis of Ferroptosis-Related Biomarker Signatures to Improve the Diagnosis and Prognosis Prediction of Ovarian Cancer. Front Cell Dev Biol 2022;9:807862. [Crossref] [PubMed]
- Shan X, Zhang C, Wang Z, et al. Prognostic value of a nine-gene signature in glioma patients based on tumor-associated macrophages expression profiling. Clin Immunol 2020;216:108430. [Crossref] [PubMed]
- Yan H, Qu J, Cao W, et al. Identification of prognostic genes in the acute myeloid leukemia immune microenvironment based on TCGA data analysis. Cancer Immunol Immunother 2019;68:1971-8. [Crossref] [PubMed]
- Daver N, Garcia-Manero G, Basu S, et al. Efficacy, Safety, and Biomarkers of Response to Azacitidine and Nivolumab in Relapsed/Refractory Acute Myeloid Leukemia: A Nonrandomized, Open-Label, Phase II Study. Cancer Discov 2019;9:370-83. [Crossref] [PubMed]
- Devarakonda S, Rotolo F, Tsao MS, et al. Tumor Mutation Burden as a Biomarker in Resected Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:2995-3006. [Crossref] [PubMed]
- Lee DW, Han SW, Bae JM, et al. Tumor Mutation Burden and Prognosis in Patients with Colorectal Cancer Treated with Adjuvant Fluoropyrimidine and Oxaliplatin. Clin Cancer Res 2019;25:6141-7. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Chen EX, Jonker DJ, Loree JM, et al. Effect of Combined Immune Checkpoint Inhibition vs Best Supportive Care Alone in Patients With Advanced Colorectal Cancer: The Canadian Cancer Trials Group CO.26 Study. JAMA Oncol 2020;6:831-8. [Crossref] [PubMed]
- Du J, Wang T, Li Y, et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radic Biol Med 2019;131:356-69. [Crossref] [PubMed]
- Yusuf RZ, Saez B, Sharda A, et al. Aldehyde dehydrogenase 3a2 protects AML cells from oxidative death and the synthetic lethality of ferroptosis inducers. Blood 2020;136:1303-16. [Crossref] [PubMed]
- Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther 2020;5:108. [Crossref] [PubMed]
- Hakimi AA, Reznik E, Lee CH, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016;29:104-16. [Crossref] [PubMed]
- Haroun F, Solola SA, Nassereddine S, et al. PD-1 signaling and inhibition in AML and MDS. Ann Hematol 2017;96:1441-8. [Crossref] [PubMed]
- Bruserud Ø, Hovland R, Wergeland L, et al. Flt3-mediated signaling in human acute myelogenous leukemia (AML) blasts: a functional characterization of Flt3-ligand effects in AML cell populations with and without genetic Flt3 abnormalities. Haematologica 2003;88:416-28. [PubMed]


