
Upregulated YTHDF1 associates with tumor immune microenvironment in head and neck squamous cell carcinomas
Introduction
As the sixth malignant tumor worldwide, head and neck squamous cell carcinomas (HNSCC) has a roughly 40–50% 5-year overall survival rate (1,2). Despite surgery, chemotherapy and radiotherapy being performed, the HNSCC patients’ overall survival rate also stays at a low level (3,4). In most HNSCC patients, metastatic and recurrent events frequently occur and are currently incurable, which becoming increasingly urgent for more effective therapies. Compared to traditional treatments like surgery, radiotherapy and chemotherapy, immunotherapy achieved better outcomes for patients (5-7). Although immunotherapy employed Anti-PD-1/L1 and Anti-CTLA-4 in possession of considerable clinical effects for HNSCC patients, there are still some patients has immune resistance (8,9). The complex tumor immune microenvironment (TME) contained stromal cells, distant recruited cells, secreted factors (such as chemokines, cytokines, and growth factors), and new blood vessels. TME components interact with tumor cells and drive tumor cell biological behavior, inducing proliferation, angiogenesis, and immune tolerance (10,11). Notably, the characteristic of TME cell infiltration is considered an anti-cancer immune response predictive factor, which increases the success of immunotherapy (11-13). Therefore, determining the TME characteristics and finding potential biomarkers will be significant for new therapeutic methods.
N6-methyladenosine (m6A) is regulated by the methyltransferases (also known as writers), demethylases (also known as erasers) and binding proteins (also known as readers), which involved in RNA splicing, mRNA formation and stability (14-17). In the tumor cells, the m6A regulators have crucial roles in tumor cell proliferation, energy metabolism, and immunomodulatory abnormalities (18-20). Specifically, different expressions of m6A regulators associate with variant activities of tumor hallmark-related pathways, which lead to tumorigenesis and tumor progression (18,19). In immune cells, m6A modification is responsible for the series of signal events that result in tumor immunity (21,22). For example, abnormalities of m6A mRNA in dendritic cells powerfully contributed to immune disorders and tumor immune escape (22). Moreover, m6A mRNA methylation also has an impact on extracellular signaling stimulations (including cytokines, chemokines, and high-pressure physical stresses), which affect various aspects of tumor progression (23). Although increasing evidence indicates that the aberrant m6A modifications are necessary for HNSCC progression, whether m6A regulators involve in HNSCC TME infiltration and which one is most relevant to TME is not yet known.
As an m6A reader, YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) belongs to the YTH domain family. YTHDF1 regulates mRNA stability by promoting the degradation of m6A-containing mRNAs (24). Although previous studies proved that YTHDF1 plays a crucial role in cancer development and durable neoantigen-specific antitumor immunity (22,25), the roles of YTHDF1 in HNSCC TME are not clear. Here, a computational screen was applied to identify m6A regulators that are most relevant to TME. Among the 24 m6A regulators, we systematically identified YDHDF1 as a potentially critical factor of HNSCC TME. Focused on YTHDF1, we found that YTHDF1 was up-regulated and amplified in HNSCC, which might be activated by SOX2 and TP63. Moreover, YTHDF1 expression positively correlates with infiltration of CD4+ T cells, while negatively correlates with the cancer-immunity cycle and immune checkpoint inhibitors. The TME infiltration characteristics and underlying mechanism of YTHDF1 in HNSCC might be critical for guiding immunotherapy and improving prognosis. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-503/coif).
Methods
The collection of data
The data that consist of mRNA expression, DNA methylation and clinical annotation data were downloaded from The Cancer Genome Atlas (TCGA) (26). The somatic mutation and copy number variation (CNV) data of HNSCC were downloaded from the Xena and cBioPortal (26,27). Infiltration Estimation of TCGA HNSCC patients were downloaded from TIMER2.0 (28,29). Immune activity scores on TCGA HNSCC samples downloaded from Tumor Immunophenotype Profiling (TIP) (30). The HNSCC assay for transposase-accessible chromatin with high throughput sequencing (ATAC-Seq) data were obtained from the Genomic Data Commons (GDC) (31). The HNSCC cell lines H3K27ac, SOX2 and TP63 chromatin immunoprecipitation followed by sequencing (ChIP-seq) data were obtained from GSE88976 and GSE104137 (32-34).
Identification of immune-related m6A regulators
To discover immune-related m6A regulators, 24 m6A regulators were analyzed, including 14 readers, 8 writers and 2 erasers (35-38). For m6A regulators differential expression, 44 normal and 520 primary tumor tissues that achieved mRNA expression profiles were used. Differentially expressed m6A regulators in the various groups were calculated by R package (39). Winn diagram was performed for differentially expressed m6A regulators, whose P value <0.01.
Somatic mutation, CNV analysis
In accordance with the TCGA publication guidelines (40,41), the mutation analysis was performed using cBioPortal for Cancer Genomics data based on the study of HNSCC (TCGA, Pan-Cancer Atlas) (27). Using the mean cut-offs of the GISTIC algorithm (40), genes were categorized in different groups, including deletion, diploid, gain and amplification. The frequency of CNV was calculated using the formula: (amplification or deletion sample number) / (total number of samples).
TME infiltration and cancer-immunity cycle analysis
We performed TIMER for TME infiltration analysis, which including CD4+ T cells, B cells, CD8+ T cells, etc. (28). The Spearman’s coefficient was used for the correlation analysis. Besides, the immune cells abundance was explored according to YTHDF1 copy number. The cancer-immunity cycle is a critical frame structure for immunotherapy, which involved a battery of continuous events including (Step 1) release of cancer cell antigens, (Step 2) cancer antigen presentation, (Step 3) priming and activation, (Step 4) trafficking of immune cells to tumors, (Step 5) infiltration of immune cells into tumors, (Step 6) recognition of cancer cells by T cells and (Step 7) killing of cancer cells (30). We analyzed the correlations of YTHDF1 expression with cancer-immunity cycle by the R software with the Pearson coefficient.
Survival analysis
The KM Plotter was used for the prognostic value of YTHDF1 in HNSCC. Using an auto select best cutoff of YTHDF1 expression, the patients were separated into two groups and then the overall survival (OS) was calculated. For restrict analysis based on cellular content, several immune cells were involved, which HNSCC patients were re-organized into two groups.
Pathway enrichment analysis
To identify the interacting proteins of YTHDF1, we performed a protein-protein interaction (PPI) network analysis by GeneMANIA. For pathway enrichment analysis, differentially expressed genes (DEGs) were identified with |log (fold change)|>0.5 and P value <0.01. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) that consist of Biological Process, Cellular Component and Molecular Function were executed by The Database for Annotation, Visualization and Integrated Discovery (DAVID) (42,43).
Statistical analysis
R software 3.6, SPSS 25.0 and GraphPad Prism 6.0 were used for statistical analyses. The continuous variables were calculated by t-test (2-tailed) or Mann-Whitney U test. The Pearson or Spearman correlation coefficient was used for correlation analysis. Survival curves were constructed using Kaplan–Meier methodology. All statistical tests that P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
YTHDF1 is an immune-related m6A regulator
To search immune-related m6A regulators, a computational screen was applied to 24 m6A regulators (including 14 readers, 8 writers and 2 erasers) in TCGA HNSCC samples (Figure 1A). Notably, m6A regulators expression was frequently dysregulated in HNSCC (Figure 1A). Specifically, more than 80% (20/24) m6A regulators were differentially expressed in HNSCC, with 12 m6A regulators (YTHDF1, IGF2BP1/2/3, HNRNPA2B1, etc.) up-regulated and 8 m6A regulators (YTHDC1/2, METTL14, RBM15B, etc.) down-regulated in tumors. Interestingly, only four m6A regulators’ expression was not significantly different in HNSCC, including YTHDF3, CBLL1, FTO and ALKBH5. These results suggest that m6A regulators may involve in HNSCC progression.
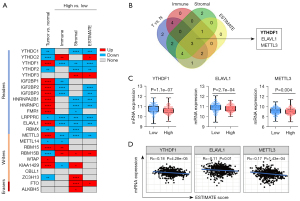
Containing complex components including immune cells, stromal cells and other factors, TME extremely affects tumor cell proliferation, angiogenesis and immune tolerance (10,11). Immune score, Stromal score and ESTIMATE score calculated by TIMER commendably reflects the immune cell infiltration, stromal components, and general situation, respectively (28). To evaluate whether m6A regulators is an immune-related factor, differential expression analysis was performed. Among 24 m6A regulators, most of m6A regulators was frequently dysregulated in their respective groups (Figure 1A). Winn Diagram showed that only YTHDF1, ELAVL1 and METTL3 were differentially expressed in each group, which may be associated with TME (Figure 1B). Corresponding to the high ESTIMATE score group, the YTHDF1, ELAVL1 and METTL3 had higher gene expression in the low group (Figure 1C). Furthermore, we found that YTHDF1, ELAVL1 and METTL3 mRNA levels highly associates with ESTIMATE score (Figure 1D). These results suggest that YTHDF1, ELAVL1 and METTL3 likely involved in the formation and alteration of TME in HNSCC. Notably, YTHDF1 expression was the most significant difference between the two groups based on the median ESTIMATE score (a score that reflects the general condition of TME) and YTHDF1 is the most correlated with the ESTIMATE score (Figure 1C,1D). Therefore, we focused on YTHDF1 for further investigations in this study.
YTHDF1 is up-regulated and amplified in HNSCC patients
As the top candidate, YTHDF1 was highly expressed in HNSCC (Figure 2A). In spite of without statistically significant, high YTHDF1 expressed patients had a shorter survival time (Figure 2B). Notably, a receiver operating characteristic (ROC) curve showed that YTHDF1 expression is sensitive and specific for diagnosis of HNSCC with the 0.74 of area under curve (AUC) (Figure 2C). Due to somatic mutation and CNVs involved in gene expression, we first analyzed the mutations of YTHDF1 in HNSCC. However, less than 0.8% of mutations of the YTHDF1 core protein domains was occurred in HNSCC patients (Figure 2D). Interestingly, we observed copy number amplification at YTHDF1 locus in HNSCC (Figure 2E). As anticipated, the mRNA expression of YTHDF1 increased in YTHDF1-amplified samples compared with diploid samples (Figure 2F). Moreover, YTHDF1 expression and its copy number has a significant positive correlation in HNSCC samples (Figure 2G). YTHDF1 copy number significantly correlates with disease specific survival (Figure 2H). These results suggested that copy-number contribute to the overexpression of YTHDF1.
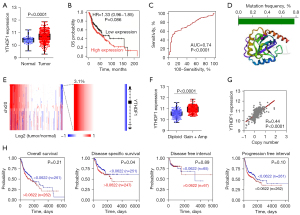
Besides genetic alterations (somatic mutation and CNV), epigenetic modifications (such as DNA methylation) also affect gene expression (44,45). Using TCGA HNSCC DNA methylation data (HumanMethylation450), we analyzed the methylation level of YTHDF1. Unfortunately, there was no difference between tumor and normal samples of the methylation level at YTHDF1 promoter region (Figure S1). This suggests that DNA methylation is not involved in regulating YTHDF1 gene expression in HNSCC.
SOX2/TP63 may activate YTHDF1 promoter in HNSCC
In addition to genetic alterations and DNA methylation, a transcriptional regulatory process based on tissue-specific transcription factors strictly regulate gene expression in HNSCC. Therefore, we attempted to reveal the molecular basis of YTHDF1 transcriptional activation in HNSCC. Using the TCGA ATAC-Seq data (31), we found that YTHDF1 promoter region exhibited high chromatin accessibility in HNSCC samples (top 3 red tracks, Figure 3A). Furthermore, the ChIP-Seq data of HNSCC cell lines showed that the promoter region of YTHDF1 had coincident H3K27ac signals (tracks 4-10, Figure 3A), suggesting a potential transcriptional activation of promoter region.
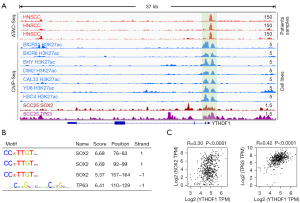
Notably, the ChIP-Seq data showed that SOX2 and TP63 had readily discernible signals at the YTHDF1 promoter region (bottom 2 tracks, Figure 3A). Therefore, it is reasonably to suspect that SOX2 and TP63 could regulate YTHDF1 expression. Indeed, Motif analysis showed that there are many binding sites of SOX2 and TP63 in the YTHDF1 promoter region (Figure 3B). Importantly, SOX2 and TP63 strongly correlates with YTHDF1 expression in HNSCC samples (Figure 3C). These results suggest that SOX2 and TP63 probably could upregulate YTHDF1 expression in HNSCC by activating its promoter.
YTHDF1 associated with cell cycle, lymphatic metastasis and angiogenesis
The expression of several kinds of tumor hallmarks was analyzed for YTHDF1 biological function, including cell proliferation, cell cycle and epithelial mesenchymal transition (EMT) regulators, stemness, lymphatic metastasis and angiogenesis. Compared with the high YTHDF1 expression group, the expression of cell cycle regulators of the low expression group was more frequently dysregulated, including 5 down-regulated cell cycle promoting factors (CCNA1, CCND1, CCNE1, MYC, and CDK6) and 3 up-regulated cell cycle inhibitors (CDKN2A, CDKN2C, CDKN1B) (Figure 4A), suggesting that YTHDF1 potentially regulate cell cycle. Moreover, tumor malignant hallmarks including CD44, SNAI2, HAS1, HAS3, EFNB2, etc. were significantly down-regulated in low YTHDF1 expression group (Figure 4A).
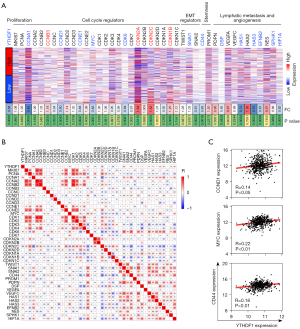
Consistently, YTHDF1 was positively correlated with these cell cycle promoting factors (CCNA1, CCND1, MYC, CDK6), EMT regulators (SNAI1), stemness (CD44), lymphatic metastasis and angiogenesis (VEGFC, HAS1, HAS2, HAS3, EFNB2, etc.) (Figure 4B,4C). The negative correlation between YTHDF1 and cell cycle inhibitors were also observed in HNSCC (Figure 4B). The implication of these results is that YTHDF1 associated with cell cycle, lymphatic metastasis and angiogenesis.
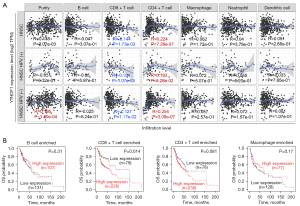
YTHDF1 correlates with CD8+ T cell infiltration
To investigate the role of YTHDF1 in infiltrating immune cells, a correlation analysis was carried out. As a result, YTHDF1 expression positively correlates with infiltration of CD4+ T cells and negatively correlates with CD8+ T cells in HNSCC (Figure 5A). Notably, the YTHDF1 copy number gain group has lower levels of most immune cells than the diploid group (Figure S2). Furthermore, high YTHDF1 expressed HNSCC patients with enriched CD8+ T cells had a poor prognosis (Figure 5B). However, YTHDF1 expression had no significant correlation with the prognosis of HNSCC, who enriched B cells, Macrophage, and CD4+ T cells (Figure 5B). These results prompt that YTHDF1 may affect HNSCC progress partly due to immune infiltration.
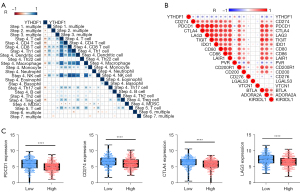
YTHDF1 negatively correlated with the cancer-immunity cycle and immune checkpoint inhibitors
Anti-cancer immune response with a battery of gradual events was considered as the cancer-immunity cycle (30), whose activities are a direct and comprehensive performance for chemokine system and other immunomodulators (30,46). Therefore, a correlation between YTHDF1 expression and cancer-immunity cycle activities was performed. Importantly, YTHDF1 expression negatively correlates with several the cancer-immunity cycle steps (Figure 6A). The above findings provided a potential role of YTHDF1 for immunosurveillance evasion in HNSCC. Furthermore, YTHDF1 negatively correlates with immune checkpoint inhibitors (such as PD-1, PD-L1, LAG-3, CTLA-4, TIGIT, IDO1, CD80, CD86, etc.) (Figure 6B). Consistently, the YTHDF1 high expression group had a lower expression of these immune checkpoint inhibitors than the YTHDF1 low expression group (Figure 6C), suggesting YTHDF1 is a potential index for quantifying the TME in HNSCC.
YTHDF1 involved in many immune-related pathways
We first performed an analysis of interaction proteins with YTHDF1 by GeneMANIA. The result showed that YTHDF1 could interact with YTHDF2/3, YTHDC1, CDKN1A, ELF1/2, RPA1/2/3, etc. (Figure 7A). The majority of these interaction proteins had a co-expression and co-localization with YTHDF1 (Figure 7A), indicating that YTHDF1 could form a complex with interaction proteins to regulate downstream gene expression.
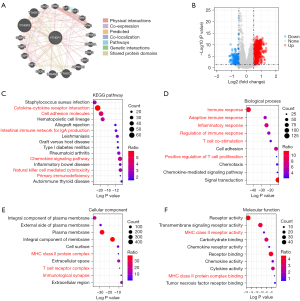
Then, we further investigated the downstream genes of YTHDF1 and found a total of 1,178 DEGs in HNSCC (Figure 7B). KEGG pathway analyses of 1,178 DEGs showed that YTHDF1-regulated genes were significantly enriched in many immune-related pathways, including cytokine-cytokine receptor interaction, cell adhesion molecules, intestinal immune network for IgA production, chemokine signaling pathway, natural killer cell mediated cytotoxicity and primary immunodeficiency, etc. (P<10e-10, Figure 7C). Furthermore, in terms of top 10 biological processes, 6 biological processes are highly related to immune response, including immune response, adaptive immune response, inflammatory response, regulation of immune response, and T cell co-stimulation (P<10e-10, Figure 7D). Surprisingly, the MHC class II protein complex was significantly enriched in both cellular component and molecular function (Figure 7E,7F). Moreover, the T cell receptor complex and immunological synapse were also enriched in cellular component (Figure 7E). These results suggest that YTHDF1 involved in many immune-related pathways in HNSCC.
Discussion
As the most commonly RNA modification, m6A participates in various human tumors progression by striking disordered RNA metabolism of downstream genes (38,47). However, the m6A regulators that is most relevant to TME in HNSCC is unknown. Here, we performed a computational screen to TCGA HNSCC patient samples and identified that YTHDF1, a specific m6A reader, were highly correlated with TME infiltration. We also revealed the molecular characteristics of YTHDF1 in HNSCC, including somatic mutation, CNVs, DNA methylation, and transcriptional regulation. Moreover, the underlying molecular mechanism of YTHDF1 in HNSCC was identified, which might be critical for guiding immunotherapy and improving prognosis.
Generally, both genetic and epigenetic variations involved in regulating gene expression. Specifically, somatic mutations and DNA hypermethylation results in gene suppression, while copy number gains could induce the gene activation (48-50). For instance, in our and other previous studies, many genes suppressed by somatic mutation and deletion were identified in squamous cell carcinomas, such as EP300, CUL3, ZNF750, etc. (26,48-50). We also have identified several genes (such as DLX5 and CEBPG) activated by copy number gains (51,52). In this study, a recurrent copy number gains of YTHDF1 were identified in HNSCC, suggesting copy number gains result in the YTHDF1 overexpression. However, somatic mutations on YTHDF1 occurred rarely in HNSCC, with a mutation frequency of only 0.8%. Moreover, the DNA methylation levels at the YTHDF1 promoter region was not significantly difference between normal and tumor tissues, suggesting that DNA methylation did not involve in regulating YTHDF1 gene expression in HNSCC.
Except for genetic and epigenetic variations, transcriptional regulation triggered by tissue-specific transcription factors tightly controls gene expression in human tumors (53). In squamous cell carcinoma, tissue-specific transcription factors SOX2 and TP63 have been well illustrated, which contribute greatly to squamous cell carcinoma progression (54,55). For instance, SOX2 and TP63 are specifically expressed in squamous cell carcinoma and form a well-organized transcriptional regulatory network to promote squamous cell carcinoma tumorigenesis, which activate the promoters, enhancers or/and super-enhancers of downstream gene. However, it is not clear whether SOX2/TP63 is involved in regulating m6A regulators gene expression. Here, we firstly found that the squamous-specific transcription factor SOX2/TP63 could directly interact with the promoter region of YTHDF1. SOX2/TP63 possibly evicts Polycomb repressive complex and recruits additional cofactors at YTHDF1 promoter, which eventually lead to YTHDF1 transcription activation. Indeed, a high chromatin accessibility of YTHDF1 promoter region was exhibited in HNSCC samples and this facilitated the recruitment of additional cofactors. The positive correlation between YTHDF1 and SOX2/TP63 expression suggest that YTHDF1 might be activated by squamous-specific transcription factor SOX2/TP63, which need to be further investigated.
In the terms of biological function, YTHDF1 promotes tumor growth in several human cancers, including the cancer of lung, ovarian, colorectal, gastric, etc. (56-58) Although Ye et al. indicated that YTHDF1 induced HNSCC cells tumorigenesis (59), its association to cell cycle and EMT regulators, stemness, lymphatic metastasis and angiogenesis, and TME infiltration remains hitherto unknown. Here, although there is not statistically correlation between YTHDF1 and cell proliferation marker (MKI67 and PCNA), we firstly identified that YTHDF1 was positively correlated with several cell cycle promoting factors, EMT regulators (SNAI1), stemness marker (CD44), lymphatic metastasis and angiogenesis and was negatively correlated with some cell cycle inhibitors. These results suggest that YTHDF1 could induce tumorigenesis and progression of HNSCC via regulating cell cycle, lymphatic metastasis and angiogenesis, which should be investigated in further research.
Recently, the effect of YTHDF1 in immunity mainly focus on innate immune response. For instance, YTHDF1 regulates innate immune response in the enteric canal (25,60) and controls CD8+ T cells in the colon cancer and melanoma (22,60). As is well-known, TME comprehensively regulate tumorigenesis, progression, metastasis and even response to immunotherapy. However, the effect of YTHDF1 on HNSCC TME is still not clear. Here, we identified that YTHDF1 positively associates with CD8+ T cell and negatively correlates with cancer-immunity cycle and immune checkpoint inhibitors. These indicated that YTHDF1 may mediates HNSCC immune evasion in multiple facets and promote tumor progression. Moreover, YTHDF1 interacts with YTHDF2/3 to regulate several immune-related pathways. Notably, most of biological process of YTHDF1 were enriched in immune-related process. It has been reported that YTHDF1 induced HNSCC tumorigenesis partially depending on iron metabolism (59), however, the relationship among complex regulation of TME, intracellular tumor iron loading and YTHDF1-induced HNSCC progression, is still required to further investigations. In the future, more studies and new techniques (including single-cell sequencing, multi-omics analysis, and nanotechnology) need to focus on the effect of YTHDF1 on the HNSCC TME, which could be a promising approach to immunotherapy.
Conclusions
In conclusion, we systematically interpreted the immune characteristics of m6A regulators in HNSCC and identified YTHDF1 as the most relevant factor of TME for HNSCC patients. As the top candidate, YTHDF1 was up-regulated and amplified in HNSCC and may transcriptionally activated by SOX2/TP63. Moreover, YTHDF1 associated with malignant phenotype and TME infiltration. Mechanistically, YTHDF1 interacts with YTHDF2/3 to regulate immune-related pathway. Thus, we provided novel insights into the YTHDF1 dysregulation in HNSCC elucidated a potential role for YTHDF1 in HNSCC TME.
Acknowledgments
We thank the researchers of previous studies for providing available data in the TCGA.
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-503/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-503/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-503/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2019;5:1749-68. [Crossref] [PubMed]
- McBride S, Sherman E, Tsai CJ, et al. Randomized Phase II Trial of Nivolumab With Stereotactic Body Radiotherapy Versus Nivolumab Alone in Metastatic Head and Neck Squamous Cell Carcinoma. J Clin Oncol 2021;39:30-7. [Crossref] [PubMed]
- Patterson RH, Fischman VG, Wasserman I, et al. Global Burden of Head and Neck Cancer: Economic Consequences, Health, and the Role of Surgery. Otolaryngol Head Neck Surg 2020;162:296-303. [Crossref] [PubMed]
- Kennedy LB, Salama AKS. A review of cancer immunotherapy toxicity. CA Cancer J Clin 2020;70:86-104. [Crossref] [PubMed]
- Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 2018;175:313-26. [Crossref] [PubMed]
- O'Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol 2019;16:151-67. [Crossref] [PubMed]
- Tang J, Shalabi A, Hubbard-Lucey VM. Comprehensive analysis of the clinical immuno-oncology landscape. Ann Oncol 2018;29:84-91. [Crossref] [PubMed]
- Boutros C, Tarhini A, Routier E, et al. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat Rev Clin Oncol 2016;13:473-86. [Crossref] [PubMed]
- McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16:717-27. [Crossref] [PubMed]
- Varn FS, Mullins DW, Arias-Pulido H, et al. Adaptive immunity programmes in breast cancer. Immunology 2017;150:25-34. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Ali HR, Chlon L, Pharoah PD, et al. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med 2016;13:e1002194. [Crossref] [PubMed]
- Alarcón CR, Goodarzi H, Lee H, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015;162:1299-308. [Crossref] [PubMed]
- Liu F, Clark W, Luo G, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016;167:816-828.e16. [Crossref] [PubMed]
- Alarcón CR, Lee H, Goodarzi H, et al. N6-methyladenosine marks primary microRNAs for processing. Nature 2015;519:482-5. [Crossref] [PubMed]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016;530:441-6. [Crossref] [PubMed]
- Li Y, Xiao J, Bai J, et al. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer 2019;18:137. [Crossref] [PubMed]
- Gu Y, Wu X, Zhang J, et al. The evolving landscape of N6-methyladenosine modification in the tumor microenvironment. Mol Ther 2021;29:1703-15. [Crossref] [PubMed]
- Chen Y, Lin Y, Shu Y, et al. Interaction between N6-methyladenosine (m6A) modification and noncoding RNAs in cancer. Mol Cancer 2020;19:94. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. [Crossref] [PubMed]
- Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell 2005;7:211-7. [Crossref] [PubMed]
- Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020;181:1582-1595.e18. [Crossref] [PubMed]
- Zong X, Xiao X, Shen B, et al. The N6-methyladenosine RNA-binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. Nucleic Acids Res 2021;49:5537-52. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [Crossref] [PubMed]
- Xu L, Deng C, Pang B, et al. TIP: A Web Server for Resolving Tumor Immunophenotype Profiling. Cancer Res 2018;78:6575-80. [Crossref] [PubMed]
- Corces MR, Granja JM, Shams S, et al. The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898.
- Zhang X, Choi PS, Francis JM, et al. Somatic Superenhancer Duplications and Hotspot Mutations Lead to Oncogenic Activation of the KLF5 Transcription Factor. Cancer Discov 2018;8:108-25. [Crossref] [PubMed]
- Sastre-Perona A, Hoang-Phou S, Leitner MC, et al. De Novo PITX1 Expression Controls Bi-Stable Transcriptional Circuits to Govern Self-Renewal and Differentiation in Squamous Cell Carcinoma. Cell Stem Cell 2019;24:390-404.e8. [Crossref] [PubMed]
- Mei S, Qin Q, Wu Q, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res 2017;45:D658-62. [Crossref] [PubMed]
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017;169:824-835.e14. [Crossref] [PubMed]
- Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. [Crossref] [PubMed]
- Tong J, Flavell RA, Li HB. RNA m6A modification and its function in diseases. Front Med 2018;12:481-9. [Crossref] [PubMed]
- Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics 2013;5:553-68. [Crossref] [PubMed]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484-92. [Crossref] [PubMed]
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1-10. [Crossref] [PubMed]
- Zhao W, Qi X, Liu L, et al. Epigenetic Regulation of m6A Modifications in Human Cancer. Mol Ther Nucleic Acids 2020;19:405-12. [Crossref] [PubMed]
- Kojima S, Cimini D. Aneuploidy and gene expression: is there dosage compensation? Epigenomics 2019;11:1827-37. [Crossref] [PubMed]
- Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: In the right place at the right time. Science 2018;361:1336-40. [Crossref] [PubMed]
- Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science 2015;349:1483-9. [Crossref] [PubMed]
- Huang Y, Lin L, Shen Z, et al. CEBPG promotes esophageal squamous cell carcinoma progression by enhancing PI3K-AKT signaling. Am J Cancer Res 2020;10:3328-44. [PubMed]
- Huang Y, Yang Q, Zheng Y, et al. Activation of bivalent factor DLX5 cooperates with master regulator TP63 to promote squamous cell carcinoma. Nucleic Acids Res 2021;49:9246-63. [Crossref] [PubMed]
- Lambert SA, Jolma A, Campitelli LF, et al. The Human Transcription Factors. Cell 2018;172:650-65. [Crossref] [PubMed]
- Watanabe H, Ma Q, Peng S, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest 2014;124:1636-45. [Crossref] [PubMed]
- Jiang Y, Jiang YY, Xie JJ, et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun 2018;9:3619. [Crossref] [PubMed]
- Pi J, Wang W, Ji M, et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res 2021;81:2651-65. [Crossref] [PubMed]
- Shi Y, Fan S, Wu M, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun 2019;10:4892. [Crossref] [PubMed]
- Liu T, Wei Q, Jin J, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res 2020;48:3816-31. [Crossref] [PubMed]
- Ye J, Wang Z, Chen X, et al. YTHDF1-enhanced iron metabolism depends on TFRC m6A methylation. Theranostics 2020;10:12072-89. [Crossref] [PubMed]
- Terajima H, Lu M, Zhang L, et al. N6-methyladenosine promotes induction of ADAR1-mediated A-to-I RNA editing to suppress aberrant antiviral innate immune responses. PLoS Biol 2021;19:e3001292. [Crossref] [PubMed]


