
Molecular basis of heterogeneity in small cell lung cancer
Introduction
Small cell lung cancer (SCLC) is an aggressive neuroendocrine differentiated neoplasm (1). Despite a high response rate to first-line cytotoxic chemotherapy, almost all patients have an early relapse. Although immune checkpoint blockade therapies for SCLC have been established recently, survival rates have not yet surpassed those of non-SCLC patients (2). Thus, novel therapeutic modalities are desirable.
During the development and treatment of tumors, including SCLC, cancer cells show and increase intratumoral heterogeneity, and acquire the resistance to therapies (3). Analysis of tumor heterogeneity could help to resolve the disease relapse. Thus, accurate assessment of tumor heterogeneity at the molecular level is crucial for the development of effective therapies (1,4).
Molecular basis and subtypes of SCLC
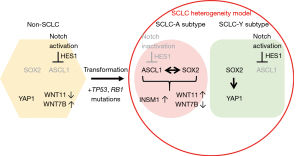
SCLC is characterized by specific gene defects, such as the TP53 and RB1 mutations found in the majority of SCLC patients. Additionally, activation of certain signaling pathways (such as PI3K/AKT) and inactivation of other pathways (such as Notch) have been demonstrated in SCLC (5,6). Comprehensive genomic analyses of human SCLC samples and cell lines have also highlighted the importance of particular transcriptional networks, as well as the molecular function of SOX2 and Achaete-Scute complex homolog 1 (ASCL1), in SCLC (7,8).
Histologically, the morphological features of SCLC, including dense sheets of small cells and scant cytoplasm, are clearly defined. In addition, for diagnosis of SCLC, the demonstration of neuroendocrine differentiation using neuroendocrine markers, including insulinoma-associated protein 1 (INSM1), is needed. However, a minority of SCLCs do not show common neuroendocrine markers. Moreover, morphological heterogeneity is evident in the combined type of SCLC, in which non-SCLC components such as adenocarcinoma, squamous cell carcinoma, or large cell carcinoma are present to differing extents (9).
Although it is difficult to distinguish between the heterogeneous components of SCLC by histopathological methods, large-scale genomic analysis of SCLC has recently begun to reveal the molecular basis of SCLC heterogeneity. For example, Rudin et al. proposed a system of nomenclature to describe SCLC subtypes according to the dominant expression of four transcription factors considered to be the master regulators of SCLC. The four subtypes were designated as (I) SCLC-A, characterized by ASCL1; (II) SCLC-N, characterized by neurogenic differentiation factor 1 (NEUROD1); (III) SCLC-Y, characterized by yes-associated protein 1 (YAP1); and (IV) SCLC-P, characterized by POU class 2 homeobox 3 (POU2F3) (10). The SCLC-A subtype is a neuroendocrine-high form of SCLC associated with high levels of ASCL1, a member of the basic helix-loop-helix family of transcription factors (11). In contrast, the SCLC-Y subtype is a neuroendocrine-low subtype of SCLC associated with the activation of NOTCH, Hippo, and RE-1 silencing transcription factor (REST) genes (12,13). ASCL1 plays a pivotal role in small cell carcinogenesis, and acts as a driver oncogene (11).
Significance of Notch signaling in SCLC
In a whole genome sequencing study, it is shown that about 25% of SCLC cases have mutations of Notch family genes (5), indicating the significance of Notch signaling in small cell lung carcinogenesis. Notch signaling is an essential cell signaling system that induces the expression of several genes, such as hairy and enhancer split-1 (Hes1) (14). Some gene transfection and knockdown experiments in SCLC cell lines demonstrated that Notch1 is involved in the suppression of cellular proliferation and neuroendocrine differentiation and enhancement of apoptosis (15,16). The Notch1-Hes1 pathway represses neuroendocrine differentiation by suppressing neuroendocrine-promoting transcription factors such as ASCL1 (17). Indeed, most classical SCLC cases and cell lines with neuroendocrine differentiation express ASCL1 and INSM1, and show an absence of NOTCH1 (18). Conversely, in non-SCLC cases, ASCL1 and/or INSM1 are negative but NOTCH1 is positive (18). Furthermore, in the combined type of SCLC, which is characterized by both SCLC and non-SCLC compartments, INSM1 is positive but NOTCH1 is negative in the SCLC compartment, while INSM1 is negative but NOTCH1 is positive in the non-SCLC compartment (19). Intriguigly, it is also reported that Notch signaling plays important roles in tumor heterogeneity of SCLC (12).
Notch-ASCL1-p53-RB axis in small cell lung carcinogenesis
A rare subset of epidermal growth factor receptor (EGFR) mutant adenocarcinomas showing resistance to treatment with EGFR tyrosine kinase inhibitors was reported to transform into SCLC (20). The transformation from adenocarcinoma to SCLC may have occurred via one of two proposed mechanisms. First, as an aspect of tumor heterogeneity, some tumor cells may have exhibited SCLC traits from the outset; second, SCLC traits may have been acquired during treatment with EGFR tyrosine kinase inhibitors. Bi-allelic inactivation of TP53 and RB1 is known to drive the formation of SCLC (5,7,10-13) and is understood to be a prerequisite for small cell lung carcinogenesis. Importantly, Niederst et al. reported that the transformation from adenocarcinoma to SCLC was always accompanied by the loss of RB1 (20). In line with these findings, Meder et al. analyzed combined type SCLC from the point of view of the NOTCH-ASCL1-p53-RB axis (21) and suggested that non-SCLC tumor tissue harboring Notch abnormalities could transform into SCLC through the acquisition of RB1 mutations. Conversely, combined SCLC could originate from pure SCLC; in this context, the non-SCLC component would be expected to show active Notch signaling and decreased levels of ASCL1/INSM1. Thus, we may speculate that the balance of Notch signaling activation and ASCL1 expression prior to mutation of RB1 and TP53 is an important factor in small cell carcinogenesis, including the formation of combined type tumors.
The role of SOX2 and Wnt signaling in SCLC
Amplification of SOX2 occurs frequently in SCLC, while suppression of SOX2 using shRNA blocks proliferation in SOX2-amplified SCLC lines (7). In addition, SOX2 has distinct effects on transcriptional programs in SCLC-A and SCLC-Y subtypes. For example, in the SCLC-A subtype, ASCL1-recruited SOX2 regulates INSM1 and WNT11 expression, whereas, in the SCLC-Y subtype, SOX2 regulates YAP1 expression (8), which, in turn, leads to the suppression of neuroendocrine differentiation (22). Furthermore, Wnt signaling also plays an important role in lung cancer cell biology. For example, WNT11 regulates neuroendocrine differentiation, cellular proliferation, and epithelial-mesenchymal transition in the SCLC-A subtype (23). ASCL1 is a key regulator of WNT11–WNT7b balance in lung cancer; SCLC cells strongly express WNT11, whereas WNT7b is frequently expressed in non-SCLC cells. In conclusion, the intratumoral heterogeneity of lung cancer could be explained partly by these distinct transcriptional programs and cell signaling systems (Figure 1).
A case of combined SCLC with enteric adenocarcinoma
Wang et al. have recently reported a case of combined SCLC with enteric adenocarcinoma (24). Their study included an assessment of the expression of two transcriptional factors, thyroid transcription factor-1 (TTF-1) and caudal type homeobox 2 transcription factor (CDX2) in this rare neoplasm. TTF-1 is known to be a useful marker for the diagnosis of thyroid and lung cancers, while CDX2 plays a critical role in intestinal development and has been widely used as a diagnostic marker of intestinal differentiation (25). These two markers have also been found to aid in the identification of tumor origin in cases where the primary lesion is unknown. Intriguingly, immunohistochemical analysis revealed expression of both TTF-1 and CDX2 proteins in the enteric adenocarcinoma component of the tumor. In addition, further defects, namely EGFR p.L861Q mutation and breast cancer susceptibility gene (BRCA2) deficiency, were detected in the biopsied tissue by next-generation sequencing. For further verification, sequencing of these genes in both SCLC and enteric adenocarcinoma components would be desirable. Detection of common gene mutations in both histological tumor types would enable us to conclude that the two components of this neoplasm originated from genetically identical cells and to hypothesize that the SCLC and enteric adenocarcinoma traits were acquired through a common process of carcinogenesis. Furthermore, analysis of the Notch pathway, as well as ASCL1, SOX2, RB1, and Wnt signaling-related genes could provide valuable information to verify the mechanism of intratumoral heterogeneity in this case.
Acknowledgments
The author thanks Sarah Ivins, PhD, from Edanz (https://jp.edanz.com/ac) and Takaaki Ito, MD, DMSc, for editing a draft of this manuscript.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2224/coif). The author has no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bunn PA Jr, Minna JD, Augustyn A, et al. Small Cell Lung Cancer: Can Recent Advances in Biology and Molecular Biology Be Translated into Improved Outcomes? J Thorac Oncol 2016;11:453-74. [Crossref] [PubMed]
- Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81-94. [Crossref] [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Liang X, Lin A, Wang Q, et al. Cell plasticity in patients with NSCLC: The controversial origins of transformed SCLC. Biomed Pharmacother 2022;149:112909. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Tenjin Y, Matsuura K, Kudoh S, et al. Distinct transcriptional programs of SOX2 in different types of small cell lung cancers. Lab Invest 2020;100:1575-88. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol 2015;10:1240-2. [Crossref] [PubMed]
- Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289-97. [Crossref] [PubMed]
- Borromeo MD, Savage TK, Kollipara RK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 2016;16:1259-72. [Crossref] [PubMed]
- Lim JS, Ibaseta A, Fischer MM, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017;545:360-4. [Crossref] [PubMed]
- Zhang W, Girard L, Zhang YA, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 2018;7:32-49. [Crossref] [PubMed]
- Rizzo P, Osipo C, Foreman K, et al. Rational targeting of Notch signaling in cancer. Oncogene 2008;27:5124-31. [Crossref] [PubMed]
- Ito T, Kudoh S, Ichimura T, et al. Small cell lung cancer, an epithelial to mesenchymal transition (EMT)-like cancer: significance of inactive Notch signaling and expression of achaete-scute complex homologue 1. Hum Cell 2017;30:1-10. [Crossref] [PubMed]
- Wael H, Yoshida R, Kudoh S, et al. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer 2014;85:131-40. [Crossref] [PubMed]
- Ito T, Udaka N, Yazawa T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 2000;127:3913-21. [Crossref] [PubMed]
- Fujino K, Motooka Y, Hassan WA, et al. Insulinoma-Associated Protein 1 Is a Crucial Regulator of Neuroendocrine Differentiation in Lung Cancer. Am J Pathol 2015;185:3164-77. [Crossref] [PubMed]
- Ito T. Intratumoral heterogeneity of Notch1 expression in small cell lung cancer. J Thorac Dis 2018;10:1272-5. [Crossref] [PubMed]
- Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377. [Crossref] [PubMed]
- Meder L, König K, Ozretić L, et al. NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas. Int J Cancer 2016;138:927-38. [Crossref] [PubMed]
- Saito H, Tenjin Y, Yamada T, et al. The role of YAP1 in small cell lung cancer. Hum Cell 2022;35:628-38. [Crossref] [PubMed]
- Tenjin Y, Kudoh S, Kubota S, et al. Ascl1-induced Wnt11 regulates neuroendocrine differentiation, cell proliferation, and E-cadherin expression in small-cell lung cancer and Wnt11 regulates small-cell lung cancer biology. Lab Invest 2019;99:1622-35. [Crossref] [PubMed]
- Wang S, Tan Y, Li L, et al. A case report of pulmonary combined small cell carcinoma with enteric adenocarcinoma. Transl Cancer Res 2022; [Crossref] [PubMed]
- Lee H, Fu Z, Koo BH, et al. The expression of TTF1, CDX2 and ISL1 in 74 poorly differentiated neuroendocrine carcinomas. Ann Diagn Pathol 2018;37:30-4. [Crossref] [PubMed]


