
Development of a novel angiogenesis-related lncRNA signature to predict the prognosis and immunotherapy of glioblastoma multiforme
Introduction
Glioma is one of the most common human primary brain tumors and has a poor prognosis (1,2). Among all gliomas, glioblastoma multiforme (GBM) is the most frequent, accounting for 55% of gliomas globally (3). Despite the standard treatment of surgery combined with radiotherapy and chemotherapy, the prognosis of patients with GBM is still unsatisfactory (2). Therefore, it is urgent to find more effective treatments for glioblastoma. Prognostic analysis can provide new ideas for treatment. However, due to the complicated heterogeneity of glioblastoma, the prognosis often varies across different patients (4). Angiogenesis levels of glioma are closely related to tumor malignancy and prognosis (5). Angiogenesis is a complex multistep biological process that can provide nutrition and oxygen to the glioblastoma, which promotes solid tumor growth and progression (6). An animal study has confirmed that inhibiting angiogenesis in a GBM animal model can inhibit tumor growth (7). Furthermore, a clinical study has also shown that angiogenesis inhibitors can improve the prognosis of patients with GBM (8). Therefore, antiangiogenic therapy has been considered a very promising treatment strategy for GBM.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that do not encode proteins (9). The abnormal expression of functional lncRNAs regulates the tumorigenesis (10), proliferation (11), development (12), and other biological behaviors of glioma. A previous study has shown a close relationship between lncRNAs and angiogenesis in glioma (13). Therefore, the study of angiogenesis-related lncRNAs can provide a theoretical basis for revealing the angiogenic mechanism of GBM.
Our study aimed to investigate the role of angiogenesis-related lncRNAs in the prognosis and immunotherapy of GBM and construct a prognostic model according to lncRNAs to predict the prognosis and treatment of GBM. We identified differentially expressed lncRNAs (DE-lncRNAs) in GBM patients and established an angiogenesis-related prognostic signature using The Cancer Genome Atlas (TCGA)-GBM cohort. Then, we estimated the prognostic value of this prognostic model. Meanwhile, a nomogram was built to predict individual survival probabilities by combining clinicopathological characteristics and a prognostic model. Furthermore, immune infiltration, immunotherapy, and drug sensitivity analyses were administered to further confirm the predictive and prognostic value of the prognostic model. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1592/rc).
Methods
Data collection
Transcriptome sequencing (3 levels) data and corresponding clinical information of the TCGA-GBM cohort, including 158 GBM samples with fully available survival data, 11 GBM samples with unavailable survival data, and 5 normal tissue samples, were obtained from TCGA database (https://tcga-data.nci.nih.gov/tcga/) (Table S1). Moreover, transcriptome sequencing data of 133 GBM samples with fully available survival data in the mRNAseq-693 dataset were downloaded and used as a validation cohort from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/) (Table S2). Additionally, a total of 48 angiogenesis-related genes were acquired from the gene set enrichment analysis (GSEA)-Molecular Signatures Database v7.4 (http://www.gseamsigdb.org/gsea/msigdb/index.jsp).
Screening of angiogenesis-related differentially expressed lncRNAs (AR-DElncRNAs)
Using the “limma” package (version 3.44.3) in R (The R Foundation for Statistical Computing, Vienna, Austria), the DE-lncRNAs were identified between 5 normal and 169 tumor tissues in the TCGA-GBM cohort based on adjusted standards of P<0.05 and |log2 (fold change)| >1. The “ggplot” package (version 3.3.3) was used to plot the volcano plot. The Spearman test was then used to perform a correlation analysis between 48 angiogenesis-related genes and lncRNAs of the GBM expression matrix to screen angiogenesis-related lncRNAs. The screening criteria were |cor| >0.3 and P<0.01. Then, the angiogenesis-related lncRNAs were intersected with the DE-lncRNAs as AR-DElncRNAs.
Construction and validation of an angiogenesis-related lncRNA signature
First, GBM samples in the TCGA-GBM cohort were randomly divided into a training set (n=111) and an internal validation set (n=47) at a ratio of 7:3. The clinicopathologic characteristic of patients with GBM in the training set, validation set, and CGGA_693 cohort are shown in Table 1. Following this, a univariate Cox regression analysis was exploited to select overall survival (OS)-associated lncRNAs in the training set (P<0.05). To further narrow down the candidate lncRNAs, we applied the least absolute shrinkage and selection operator (LASSO) algorithm to prevent model overfitting by using the “glmnet” package (version 4.1-1) in R.
Table 1
| Characteristic | TCGA-GBM | CGGA_693 (n=133) | ||
|---|---|---|---|---|
| Total (n=158) | Training set (n=111) | Testing set (n=47) | ||
| Age (years), mean (± SD) | 59.6 (±13.8) | 59.0 (±12.9) | 61.0 (±15.7) | 52.3 (±13.2) |
| Gender, n (%) | ||||
| Female | 52 (32.9) | 36 (32.4) | 16 (34.0) | 53 (39.8) |
| Male | 95 (60.1) | 65 (58.6) | 30 (63.8) | 80 (60.2) |
| Unclear | 11 (7.0) | 10 (9.0) | 1 (2.1) | – |
| Vital, n (%) | ||||
| Alive | 29 (18.4) | 19 (17.1) | 10 (21.3) | 23 (17.3) |
| Dead | 128 (81.0) | 91 (82.0) | 37 (78.7) | 110 (82.7) |
| Unclear | 1 (0.6) | 1 (0.9) | – | – |
| KPS, mean (± SD) | 75.2 (±14.2) | 75.8 (±14.2) | 74.2 (±14.4) | – |
| Radio_status | ||||
| Treated | 129 (81.6) | 94 (84.7) | 35 (74.5) | 110 (82.7) |
| Untreated | 20 (12.7) | 11 (9.9) | 9 (19.1) | 19 (14.3) |
| Unclear | 9 (5.7) | 6 (5.4) | 3 (6.4) | 4 (3.0) |
| Chemo_status, n (%) | ||||
| Treated | 127 (80.4) | 91 (82.0) | 36 (76.6) | 109 (82.0) |
| Untreated | 21 (13.3) | 12 (10.8) | 9 (19.1) | 19 (14.3) |
| Unclear | 10 (6.3) | 8 (7.2) | 2 (4.3) | 5 (3.7) |
| Original subtype, n (%) | ||||
| Classical | 37 (23.4) | 24 (21.6) | 13 (27.7) | – |
| G-CIMP | 8 (5.1) | 6 (5.4) | 2 (4.3) | – |
| Mesenchymal | 45 (28.5) | 29 (26.1) | 16 (34.0) | – |
| Neural | 26 (16.4) | 21 (18.9) | 5 (10.6) | – |
| Proneural | 29 (18.4) | 20 (18.0) | 9 (19.1) | – |
| Unclear | 13 (8.2) | 11 (9.9) | 2 (4.3) | – |
| IDH status, n (%) | ||||
| Mutant | 10 (6.3) | 8 (7.2) | 2 (4.2) | 21 (15.8) |
| WT | 132 (83.5) | 90 (81.1) | 42 (89.4) | 105 (78.9) |
| Unclear | 16 (10.1) | 13 (11.7) | 3 (6.4) | 7 (5.3) |
| MGMT promoter status, n (%) | ||||
| Methylated | 51 (32.3) | 34 (30.6) | 17 (36.2) | 63 (47.4) |
| Unmethylated | 67 (42.4) | 44 (39.6) | 23 (48.9) | 54 (40.6) |
| Unclear | 40 (25.3) | 33 (29.7) | 7 (14.9) | 16 (12.0) |
| 1p19q_status, n (%) | ||||
| Codel | – | – | – | 4 (3.0) |
| Noncodel | – | – | – | 104 (78.2) |
| Unclear | – | – | – | 25 (18.8) |
CGGA, Chinese Glioma Genome Atlas; Codel, codeletion; GBM, glioblastoma multiforme; G-CIMP, Glioma CpG island methylator phenotype; IDH, isocitrate dehydrogenase; KPS, Karnofsky Performance Scale; MGMT, O6-methylguanine-DNA methyltransferase; SD, standard deviation; TCGA, The Cancer Genome Atlas; WT, wild type.
A risk score was calculated by LASSO regression coefficients using the following formula:
where coef (genei) is the risk coefficient, and expr (genei) is the expression level of prognostic lncRNAs. Based on the median risk score, samples in the training set were divided into high- and low-risk groups. Kaplan-Meier survival analysis was used to determine the survival difference between these 2 risk groups. To assess the performance of the prognostic model, area under the receiver operating characteristic (ROC) curve (AUC) analysis was conducted using the “timeROC” package (version 1.0.3) in R. In addition, the risk scores of patients with GBM in both the internal validation set and external validation set were calculated using the same formula as the methods mentioned above and used to validate the performance of the risk signature separately.
Correlation analysis of the risk model and clinical characteristics
To further explore the correlation between the risk signature and clinical characteristics, we compared the risk scores among patients with GBM with different clinical characteristics in the TCGA-GBM cohort, including age (≥65 vs. <65), sex (female vs. male), O6-methylguanine-DNA methyltransferase (MGMT) status, and isocitrate dehydrogenase 1 (IDH1), and subtype. The results were visualized by drawing violin plots with the “ggpubr” package (version 0.4.0).
Independent prognostic factor analysis and nomogram construction
Univariate and multivariate Cox regression analyses were performed to investigate the prognostic significance of clinical characteristics and risk scores in the TCGA-GBM cohort. The risk score and clinicopathological factors, including age, sex, MGMT status, IDH1 status, and pathological subtypes, were used to perform univariate Cox analysis to screen prognostic factors. Moreover, prognostic factors (P<0.05) were uploaded to multivariate Cox analysis to identify independent prognostic factors. Based on the results of the multivariate analysis, we applied the “rms” package (version 6.2-0) in R to create a nomogram for guiding clinical decision-making. The calibration curve was used to assess the predictive accuracy of the nomogram.
Construction of a competing endogenous RNA (ceRNA) regulatory network of the prognostic lncRNA signature
To predict the ceRNA network of the lncRNA signature, we used the StarBase 2.0 database (https://starbase.sysu.edu.cn/starbase2/) to predict lncRNA-microRNA (miRNA) targeting relationships with a screening condition of stringency (≥1) and used the miRWalk database (http://mirwalk.umm.uni-heidelberg.de/) to predict the miRNA-messenger RNA (mRNA) relationship pairs with a screening threshold of 1. The predicted target genes were intersected with the downregulated mRNAs according to the regulatory relationship of ceRNA (the lncRNA and mRNA expression trends were the same), and genes with Pearson correlation of |cor| > 0.5 and P<0.05 were used to construct the ceRNA network. Finally, the targeting relationships between lncRNAs, miRNAs, and mRNAs were imported into Cytoscape (version 3.8.2) to construct the lncRNA-miRNA-mRNA network.
Functional enrichment analysis
To further examine the prognostic features of the functions performed by lncRNA target genes, this study used the “clusterProfiler” package (version 3.18.0) to perform an enrichment analysis based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Doing so enabled us to identify the common functions and related pathways of a large number of genes within the key gene set. The GO system consists of 3 parts: biological process, molecular functions, and cellular components. GO terms and KEGG pathways were selected if the P value was less than 0.05 and the count showed 2 or more.
Assessment of tumor immune cell infiltration
To explore immunological differences between the high- and low-risk groups, we performed Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) analysis using the “estimate” package (version 1.0.13) to obtain the tumor tissue immune score, stromal score, and ESTIMATE score of both combined. Single-sample gene set enrichment analysis (ssGSEA) was employed to analyze the differences in immune infiltration between the high- and low-risk groups. The abundance of the 24 immune cells was visualized using “ggplot2” (version 3.3.3) and “ggpubr” (version 0.4.0) to draw box line plots. In addition, the proportion of 22 immune cell species in TCGA-GBM was calculated using the CIBERSORT algorithm (version 1.03) and the LM22 gene set. The results of scoring 22 immune cell species were visualized by drawing violin plots using the “vioplot” package (version 0.3.7). In addition, we also used the EPIC, MCP-Counter, and quanTIseq methods in the “immunedeconv” package to obtain the percentage of different immune cells. The proportion of immune and nonimmune cells in the tumor microenvironment was analyzed using the online database xCell (https://xcell.ucsf.edu/). Finally, leukocyte fraction data for the GBM samples were obtained from the Genomic Data Commons (GDC; https://gdc.cancer.gov/about-data/publications/panimmune) database, and then differences between high- and low-risk groups were compared using a rank sum test.
Immunotherapy analysis
First, we compared the expression of immune checkpoint genes in the high- and low-risk groups. Then, differences in immunotherapy sensitivity between the high- and low-risk groups were assessed using Tumor Immune Dysfunction and Exclusion (TIDE). Using the submap method, we compared differences in sensitivity of GBM to different immunotherapies. According to different therapeutic targets and responses, the sensitivity was divided into programmed cell death protein 1 (PD-1)-response (R), PD-1-no response (noR), cytotoxic T-lymphocyte protein 4 (CTLA4)-R, and CTLA4-noR.
Drug susceptibility analysis
To further examine whether risk scores could be used to predict the effectiveness of chemotherapy this study used the “pRRophetic” package (version 0.5) to calculate the half maximal inhibitory concentration (IC50) of the GBM sample for drugs in the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/). The differences in IC50 of chemotherapeutic agents between the high- and low-risk groups were compared according to the calculated results.
Statistical analysis
R language was the main tool used to generate figures and perform the statistical analysis. The use of several R language packages is described above. A P value less than 0.05 was considered to indicate a statistically significant difference.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of prognostic angiogenesis-related lncRNAs in GBM
A total of 277 lncRNAs with significant expression differences in TCGA-GBM were enrolled in this study, of which 104 were upregulated and 173 were downregulated (Figure 1A). Spearman correlation analysis revealed a total of 5,681 lncRNAs associated with 48 angiogenic genes in the lncRNA expression matrix of GBM. Candidate lncRNAs were intersected with DE-lncRNAs to obtain 255 AR-DElncRNAs (Figure 1B). Univariate Cox regression analysis then identified 3 AR-DElncRNAs that were significantly associated with OS (P<0.05) (Figure 1C). The 3 most important prognosis-related lncRNAs (DGCR5, PRKAG2-AS1, and ACAP2-IT1) were further screened out using LASSO analysis (Figures 1D,1E).
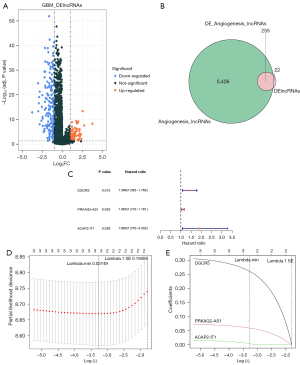
Establishment and validation of the angiogenesis-related lncRNA signature
A risk model was constructed using the expression of the 3 identified prognosis-related AR-DElncRNAs and their corresponding regression coefficients in the TCGA-GBM training set. The risk score was calculated as follows: risk score =0.31 × DGCR5 + 0.07× PRKAG2-AS1 + 0.02 × ACAP2-IT1. All patients were divided into high- and low-risk groups based on the median risk score of 0.9195 in the TCGA-GBM training set. Figure 2A shows the distribution of survival status and risk score and indicates that more deaths occurred in the high-risk group. Figure 2B displays the expression characteristics of these 3 identified prognostic signatures. High expression of the 3 prognostic signatures occurred in patients with high-risk scores. To verify the survival differences between the 2 groups, we performed survival analysis on all cases and found that the OS of patients in the high-risk group was significantly worse than that in the low-risk group (Figure 2C; P<0.05). ROC curves were plotted for patient survival from 1 to 5 years, and all AUC values were above 0.6, indicating good efficacy of the risk model (Figure 2D).
Validation sets showed better prediction accuracy of our 3 prognostic signatures. In the internal validation set TCGA-GBM, patients were classified into high- and low-risk groups according to a median risk score of 0.8113. Patients in the high-risk group were found to have a worse prognosis and higher expression of the 3 prognostic signatures than those in the low-risk group (Figures 2E-2G). The AUC values of the patients’ ROC curve analysis from 1 to 5 years were all over 0.6 (Figure 2H). Consistent results were obtained in the external validation set mRNAseq-693 (Figure S1). We plotted the unsupervised heatmap of the expression of the 3 lncRNAs (Figure 2I).
Differences in risk scores for clinical characteristics
To further investigate the prognosis of clinicopathological characteristics, the Pearson correlation between clinicopathological factors and risk score was analyzed. The correlations between the risk score and sex, age, MGMT status, and IDH1 status were not significant (P>0.05; Figures 3A-3D). Among the subtypes, the proneural subtypes had a significantly lower risk score than did the mesenchymal subtypes (P<0.05; Figure 3E).

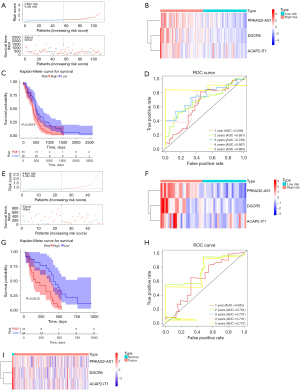
The lncRNA signature as an independent prognostic factor and construction of the nomogram
To estimate critical prognostic factors and the clinical suitability of the prognostic model, we carried out univariate and multivariate Cox analyses, from which we identified independent prognostic factors and formulated a nomogram. The results of the univariate analysis showed that risk score, age, MGMT status, and IDH1 status were statistically significant (P<0.05; Figure 4A). After the multivariate Cox analysis, we found that the risk score was a dependable independent prognostic factor for patients with GBM [hazard ratio (HR) =1.444; P=0.042; Figure 4B]. A predictive nomogram was constructed to predict the 1-, 2-, and 3-year survival rates of GBM cases based on the risk score, age, and MGMT status (Figure 4C). The concordance index of the nomogram was calculated to be 0.6742836, indicating that the model was effective in predicting 1 to 3-year survival (Figures 4D-4F).
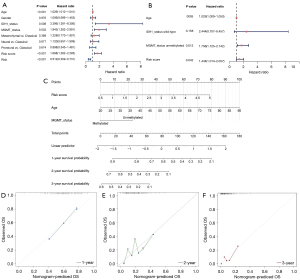
Construction of the ceRNA network of the lncRNA signature and functional analysis
To better investigate the regulatory mechanism of the lncRNA signature in GBM, we constructed a lncRNA signature-related ceRNA network. First, 29 miRNAs with targeting relationships with lncRNAs were obtained using the Starbase2.0 database (lncRNA-miRNA). Then, the target mRNAs of 29 miRNAs were predicted in the miRWalk database. According to the expression downregulation characteristics of the lncRNA signatures ACAP2-IT1, PRKAG2-AS1, and DGCR5, the predicted target genes were intersected with the differentially downregulated mRNAs. Finally, a ceRNA network containing 3 lncRNAs, 29 miRNAs, and 69 mRNAs was constructed based on the genes (|cor| >0.5; P<0.05; Figure 5A).
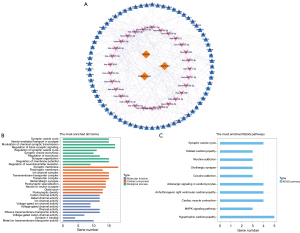
The functions of the target genes of the lncRNA signature were further analyzed. GO functional enrichment results showed that the target genes were significantly associated with biological processes, such as regulation of neurotransmitter secretion, synaptic organization, modulation of chemical synaptic transmission, regulation of membrane potential, regulation of exocytosis, synaptic vesicle exocytosis, regulation of the synaptic vesicle cycle, and regulation of trans-synaptic signaling. In terms of cellular composition, target genes were significantly related to the functions of synaptic membranes, presynaptic membranes, transport complexes, postsynaptic density, distal axons, neuron-to-neuron synapses, postsynaptic specialization, glutamatergic synapses, transmembrane transporter complexes, and ion channel complexes. In terms of molecular function, the target genes were significantly linked to voltage-gated channel activity, ion channel activity, cation channel activity, metal ion transmembrane transporter activity, syntaxin-1 binding, and passive transmembrane transporter activity (Figure 5B). KEGG analysis showed a significant correlation between target genes and myocardial contraction, hypertrophic cardiomyopathy, the mitogen-activated protein kinase (MAPK) signaling pathway, the synaptic vesicle cycle, hypertrophic cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, adrenergic signaling in cardiomyocytes, cocaine addiction, and cholinergic synapse (Figure 5C).
The lncRNA signature was associated with the immune microenvironment
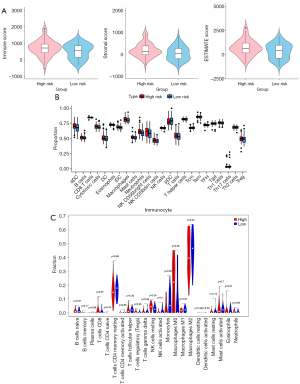
The findings of the immune microenvironment analysis revealed that the immune score, stromal score, and combined ESTIMATE score of the 2 were higher in the high-risk group than in the low-risk group, indicating high immune cell infiltration in the high-risk group (Figure 6A). According to ssGSEA, the proportion of neutrophils, macrophages, immature dendritic cells (iDCs), natural killer (NK) CD56dim cells, and activated DCs (aDCs) were significantly different between the high- and low-risk groups (P<0.05; Figure 6B). In addition, only the proportion of resting NK cells was different between the groups in the CIBERSORT algorithm results (P<0.05; Figure 6C). The results analyzed in the “immunedeconv” package showed significant differences in the proportion of macrophage M2, uncharacterized cells, and macrophage/monocyte cells between the high- and low-risk groups (P<0.05; Figures S2A-S2C). A total of 10 immune/nonimmune cells were significantly different between the high- and low-risk groups based on online database xCell analysis; the differences were in megakaryocytes, keratinocytes, plasmacytoid dendritic cells (pDCs), macrophages, M2 macrophages, pro-B cells, memory B cells, NK cells, T helper type 1 (Th1 cells), and melanocytes (P<0.05; Figure S2D). Furthermore, the leukocyte fraction was significantly higher in the high-risk group than in the low-risk group (P<0.05; Figure S2E). These results suggest a powerful correlation between the 3-lncRNA signature and the immune microenvironment.
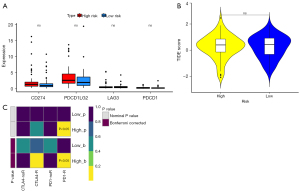
The lncRNA signature was associated with immunotherapy of PD-1-R
We assessed the correlation between the prognostic model and the expression values of immune checkpoint genes that could be used as indicators for predicting the immune response. The results demonstrated that only CD274, PDCD1LG2, LAG3, and PDCD1 immune checkpoint molecules were present in GBM samples; unfortunately, their expression did not show significant differences between the high- and low-risk groups (Figure 7A). In addition, the results of the differential assessment of immunotherapy sensitivity indicated no significant difference in the immune response in the high- and low-risk groups (Figure 7B). We further evaluated the response of the high- and low-risk groups to immunotherapy for PD-1 and CTLA4 and concluded that there was a significant difference in the sensitivity of immunotherapy for PD-1 between the high-risk group and low-risk group (Figure 7C).
The lncRNA signature could predict chemotherapy drug sensitivity
Analysis of differences in chemotherapy between high- and low-risk groups identified 28 drugs with significant differences in IC50 value, including OSI.906, cyclopamine, bosutinib, vinblastine, MG.132, cytarabine, AZD7762, A.770041, GSK269962A, FH535, ABT.888, pyrimethamine, salubrinal, lenalidomide, camptothecin, BIRB.0796, AS601245, NSC.87877, AICAR, MS.275, tipifarnib, cisplatin, nilotinib, dasatinib, KIN001.135, JNJ.26854165, axitinib, and A.443654. This result implies that these drugs may be potential chemotherapeutic agents for GBM (Figure 8).

Discussion
In recent years, the role of lncRNAs in the tumorigenesis and development of glioma has been gradually recognized. The function of lncRNAs is complicated and can be roughly divided into the following aspects: regulating the function of target proteins directly, regulating the stability and translation of long-stranded RNA molecules, affecting the inhibitory function of miRNAs, and regulating gene transcription (14). Many studies have shown that lncRNAs participate in the regulation of angiogenesis in glioma. Some lncRNAs can promote angiogenesis in glioma. lncRNA H19 promotes glioma angiogenesis via the miR-342-Wnt5a-beta-catenin axis (13). The lncRNA RPL34-AS1 promotes glioma angiogenesis by regulating the vascular endothelial growth factor A (VEGFA) signaling pathway (15). Other lncRNAs that have been reported to promote glioma angiogenesis include lncRNA PVT1 (16), lncRNA CCAT2 (17), and lncRNA NKILA (18).
Meanwhile, some lncRNAs show inhibitory effects on the angiogenesis of glioma. LncRNA SLC26A4-AS1 inhibits glioma angiogenesis by upregulating NPTX1 via nuclear factor kappa B subunit 1 (NFKB1) transcription factor (19). Since lncRNAs play an important role in the angiogenesis of glioma, they are considered potential targets for glioma therapy. It has been reported that the knockdown of lncRNA H19 can inhibit the proliferation, migration, and angiogenesis of glioma cells (13). Similar results have also been shown in other studies (15,17). In addition to participating in the regulation of glioma angiogenesis, lncRNAs are also related to the prognosis of patients with glioma. It has been shown that lncRNAs PVT1 and HAR1A can be used as prognostic biomarkers to indicate therapy outcomes for diffuse glioma patients (20). Some researchers constructed risk models based on immune-related lncRNAs. The results showed that the lncRNA-based risk model could be used to evaluate the prognosis of patients with glioma and predict the efficacy of immunotherapy (21).
In the present study, 3 AR-DElncRNAs (DGCR5, PRKAG2-AS1, and ACAP2-IT1) that significantly associated with the prognosis of patients with GBM were identified. lncRNA DGCR5 has been recognized as a potential tumor progression regulator. Abnormal expression of DGCR5 regulates the progression of digestive cancers by affecting cancer cell proliferation, aggression, metastasis, and drug resistance (22). In addition, DGCR5 also plays an important role in glioma. Some studies have shown that DGCR5 is significantly associated with the prognosis of patients with glioma and participates in the regulation of the immune response, immune infiltration, and cell proliferation of glioma (23,24). LncRNA PRKAG2-AS1 was reported to be a prognosis-related factor in patients with hepatocellular carcinoma (HCC) (25). Targeting PRKAG2-AS1 can significantly inhibit proliferation, migration, and invasion in HCC cells (25). LncRNA ACAP2-IT1 seems to be related to the regulation of N6-methyladenosine, which plays an important role in carcinogenesis and cancer inhibition (26). According to the results of the present study, DGCR5, PRKAG2-AS1, and ACAP2-IT1 are angiogenesis-related and are significantly associated with the OS of patients with GBM. This finding suggests that these 3 lncRNAs may provide potential therapeutic targets for further research on the antiangiogenic therapy of GBM.
We further established a risk model based on the 3 identified AR-DElncRNAs (DGCR5, PRKAG2-AS1, and ACAP2-IT1) and validated it. The results showed the good efficacy of the risk model. We then used the risk model to predict the prognosis of GBM with different clinicopathological characteristics. The results showed that the proneural subtypes had a significantly lower risk score than did the mesenchymal subtypes. The proneural subtype GBM has neuronal differentiation, which is common in young adults. The molecular pathological features of proneural GBM are IDH, TP53 mutations, and positivity for the glioma CpG island methylator phenotype (G-CIMP) and normal epidermal growth factor receptor (EGFR), PTEN, and Notch signaling. In contrast, mesenchymal GBM has mesenchymal differentiation, which is common in older adults. The molecular pathological features of mesenchymal GBM are abnormal EGFR amplification, PTEN loss, NF1 mutations, and Akt signaling (27). Compared with the mesenchymal subtype, the outcome of the proneural subtype is better (27), which is consistent with the risk score and proves the accuracy of the risk model.
To further investigate the regulatory mechanism of the lncRNA signature in GBM, we constructed a lncRNA signature–related ceRNA network, and 29 miRNAs were involved in this network. Among these miRNAs, miR-22-3p, miR-141-3p, miR-206, miR-30a-5p, miR-30b-5p, miR-491-5p, miR-655-3p, and miR-944 have been confirmed to be closely related to the progression, angiogenesis, radioresistance, and chemoresistance of glioma (28-35). Ten lncRNA signature-related pathways were identified using pathway enrichment analysis. Among them, the MAPK pathway has been confirmed to be closely related to angiogenesis, invasion, proliferation, and migration of glioma (36-38). These results indirectly link these AR-DElncRNAs to angiogenesis. However, direct evidence of the involvement of these lncRNAs in angiogenesis regulation is still lacking. The effects of these lncRNAs, miRNAs, and their target genes in the ceRNA network on glioma need to be further studied.
The GBM microenvironment contains infiltrating and resident immune cells, such as microglia, peripheral macrophages, myeloid-derived suppressor cells (MDSCs), leukocytes, CD4+ T cells, and T regulator cells (Tregs), which have a crucial role in glioma growth, metastasis, and response to treatment (39). In the present study, although the results of various analysis methods were different in the types of immune cells, in general, the immune cell infiltration in the high-risk group was higher than that of the low-risk group. Notably, the CIBERSORT algorithm results showed that the proportion of NK resting cells was higher in the high-risk group than in the low-risk group. Some studies have confirmed that infiltrating NK cells in glioma tissues are nonfunctional, possibly due to contact with immunosuppressive cells, such as glioma-associated microglia, macrophages (GAMs), MDSCs, and Tregs (39,40). These cells inhibit the activities of NK cells by suppressing NKG2D expression and the production of interferon gamma (INF-γ) (39). Meanwhile, the proportion of macrophages was higher in the high-risk group than in the low-risk group. Macrophages and microglia are the predominant immune population in gliomas and can constitute up to 30–50% of the total cellular composition (41). GAMs have been shown to engage in reciprocal interactions with neoplastic tumor cells to promote tumor growth and progression (42). The number of GAMs is higher in high-grade than in low-grade glioma and is generally a negative prognostic factor for survival (41). These results suggest that the risk model can help evaluate the tumor immune microenvironment of patients with GBM.
Our results showed that the high-risk group possessed a higher sensitivity to PD-1-R immunotherapy than did the low-risk group. Tumor-associated macrophages (TAMs) express PD-1. The expression of PD-1 on TAMs increases with tumor progression and correlates negatively with phagocytic activity against tumor cells. Blockade of PD-1/programmed death-ligand 1 (PD-L1) in vivo reduces tumor growth and increases survival in mouse models of cancer (43). Considering the higher proportion of macrophages in the high-risk group, this may be one of the mechanisms by which the high-risk group has a higher sensitivity to PD-1R immunotherapy compared to the low-risk group.
Finally, we analyzed the differences in chemotherapy drug sensitivity between the high- and low-risk groups. These results may provide valuable information for drug selection during the chemotherapy of GBM. However, the effectiveness of these drugs for GBM warrants further basic and clinical study validation.
There are a few limitations in our research. Our predictions and validation were conducted using bioinformatics technologies, and we did not conduct clinical research with our patient tissue samples. In addition, further experiments in vivo and in vitro were absent, which should be addressed in our future research. Despite these limitations, the results in this study were accurate and acquired after extensive data analysis. Our results provide a new research direction that can progress our understanding of the mechanism of glioblastoma.
In conclusion, we developed and validated an angiogenesis-related lncRNA signature for predicting the prognosis of patients with GBM. Moreover, the novel signature could be applied for therapeutic response prediction during the treatment of these patients.
Acknowledgments
Funding: This study was funded by the Key Research and Development Plan of Shaanxi Province, China (No. 2021SF-298).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1592/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1592/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1592/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. [Crossref] [PubMed]
- Shergalis A, Bankhead A 3rd, Luesakul U, et al. Current Challenges and Opportunities in Treating Glioblastoma. Pharmacol Rev 2018;70:412-45. [Crossref] [PubMed]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008;359:492-507. [Crossref] [PubMed]
- Peshes-Yeloz N, Ungar L, Wohl A, et al. Role of Klotho Protein in Tumor Genesis, Cancer Progression, and Prognosis in Patients with High-Grade Glioma. World Neurosurg 2019;130:e324-32. [Crossref] [PubMed]
- Wong ML, Prawira A, Kaye AH, et al. Tumour angiogenesis: its mechanism and therapeutic implications in malignant gliomas. J Clin Neurosci 2009;16:1119-30. [Crossref] [PubMed]
- Parmar D, Apte M. Angiopoietin inhibitors: A review on targeting tumor angiogenesis. Eur J Pharmacol 2021;899:174021. [Crossref] [PubMed]
- Ahir BK, Engelhard HH, Lakka SS. Tumor Development and Angiogenesis in Adult Brain Tumor: Glioblastoma. Mol Neurobiol 2020;57:2461-78. [Crossref] [PubMed]
- Schulte JD, Aghi MK, Taylor JW. Anti-angiogenic therapies in the management of glioblastoma. Chin Clin Oncol 2021;10:37. [Crossref] [PubMed]
- Zhao W, Shan B, He D, et al. Recent Progress in Characterizing Long Noncoding RNAs in Cancer Drug Resistance. J Cancer 2019;10:6693-702. [Crossref] [PubMed]
- Zhang X, Niu W, Mu M, et al. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J Exp Clin Cancer Res 2020;39:196. [Crossref] [PubMed]
- Yang H, Chen W, Jiang G, et al. Long non-coding RNA EWSAT1 contributes to the proliferation and invasion of glioma by sponging miR-152-3p. Oncol Lett 2020;20:1846-54. [Crossref] [PubMed]
- Li B, Lu X, Ma C, et al. Long non-coding RNA NEAT1 promotes human glioma tumor progression via miR-152-3p/CCT6A pathway. Neurosci Lett 2020;732:135086. [Crossref] [PubMed]
- Zhou Q, Liu ZZ, Wu H, et al. LncRNA H19 Promotes Cell Proliferation, Migration, and Angiogenesis of Glioma by Regulating Wnt5a/β-Catenin Pathway via Targeting miR-342. Cell Mol Neurobiol 2022;42:1065-77. [Crossref] [PubMed]
- Li D, Zhang Z, Xia C, et al. Non-Coding RNAs in Glioma Microenvironment and Angiogenesis. Front Mol Neurosci 2021;14:763610. [Crossref] [PubMed]
- Zhang D, Jiang H, Ye J, et al. A novel lncRNA, RPL34-AS1, promotes proliferation and angiogenesis in glioma by regulating VEGFA. J Cancer 2021;12:6189-97. [Crossref] [PubMed]
- Bi Y, Ji J, Zhou Y. LncRNA-PVT1 indicates a poor prognosis and promotes angiogenesis via activating the HNF1B/EMT axis in glioma. J Cancer 2021;12:5732-44. [Crossref] [PubMed]
- Sun SL, Shu YG, Tao MY. LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol Cell Biochem 2020;468:69-82. [Crossref] [PubMed]
- Chen Z, Li S, Shen L, et al. NF-kappa B interacting long noncoding RNA enhances the Warburg effect and angiogenesis and is associated with decreased survival of patients with gliomas. Cell Death Dis 2020;11:323. [Crossref] [PubMed]
- Li H, Yan R, Chen W, et al. Long non coding RNA SLC26A4-AS1 exerts antiangiogenic effects in human glioma by upregulating NPTX1 via NFKB1 transcriptional factor. FEBS J 2021;288:212-28. [Crossref] [PubMed]
- Zou H, Wu LX, Yang Y, et al. lncRNAs PVT1 and HAR1A are prognosis biomarkers and indicate therapy outcome for diffuse glioma patients. Oncotarget 2017;8:78767-80. [Crossref] [PubMed]
- Wang X, Gao M, Ye J, et al. An Immune Gene-Related Five-lncRNA Signature for to Predict Glioma Prognosis. Front Genet 2020;11:612037. [Crossref] [PubMed]
- Hu C, Xu W, Wang B, et al. Critical Functions of lncRNA DGCR5 in Cancers of the Digestive System. Curr Pharm Des 2021;27:4147-51. [Crossref] [PubMed]
- Wu X, Hou P, Qiu Y, et al. Large-Scale Analysis Reveals the Specific Clinical and Immune Features of DGCR5 in Glioma. Onco Targets Ther 2020;13:7531-43. [Crossref] [PubMed]
- He Z, Long J, Yang C, et al. LncRNA DGCR5 plays a tumor-suppressive role in glioma via the miR-21/Smad7 and miR-23a/PTEN axes. Aging (Albany NY) 2020;12:20285-307. [Crossref] [PubMed]
- Ou Y, Deng Y, Wang H, et al. Targeting Antisense lncRNA PRKAG2-AS1, as a Therapeutic Target, Suppresses Malignant Behaviors of Hepatocellular Carcinoma Cells. Front Med (Lausanne) 2021;8:649279. [Crossref] [PubMed]
- Zheng J, Guo J, Cao B, et al. Identification and validation of lncRNAs involved in m6A regulation for patients with ovarian cancer. Cancer Cell Int 2021;21:363. [Crossref] [PubMed]
- Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol 2014;232:165-77. [Crossref] [PubMed]
- Wang B, Wang K, Jin T, et al. NCK1-AS1 enhances glioma cell proliferation, radioresistance and chemoresistance via miR-22-3p/IGF1R ceRNA pathway. Biomed Pharmacother 2020;129:110395. [Crossref] [PubMed]
- Yu M, Yi B, Zhou W, et al. Linc00475 promotes the progression of glioma by regulating the miR-141-3p/YAP1 axis. J Cell Mol Med 2021;25:463-72. [Crossref] [PubMed]
- Wang D, Yang T, Liu J, et al. Propofol Inhibits the Migration and Invasion of Glioma Cells by Blocking the PI3K/AKT Pathway Through miR-206/ROCK1 Axis. Onco Targets Ther 2020;13:361-70. [Crossref] [PubMed]
- Zhao P, Wang M, An J, et al. A positive feedback loop of miR-30a-5p-WWP1-NF-κB in the regulation of glioma development. Int J Biochem Cell Biol 2019;112:39-49. [Crossref] [PubMed]
- Zhang D, Liu Z, Zheng N, et al. MiR-30b-5p modulates glioma cell proliferation by direct targeting MTDH. Saudi J Biol Sci 2018;25:947-52. [Crossref] [PubMed]
- Li W, Soufiany I, Lyu X, et al. SP1-upregulated LBX2-AS1 promotes the progression of glioma by targeting the miR-491-5p/LIF axis. J Cancer 2021;12:6989-7002. [Crossref] [PubMed]
- Xin J, Zhao YH, Zhang XY, et al. LncRNA NFIA-AS2 promotes glioma progression through modulating the miR-655-3p/ZFX axis. Hum Cell 2020;33:1273-80. [Crossref] [PubMed]
- Jiang J, Lu J, Wang X, et al. Glioma stem cell-derived exosomal miR-944 reduces glioma growth and angiogenesis by inhibiting AKT/ERK signaling. Aging (Albany NY) 2021;13:19243-59. [Crossref] [PubMed]
- Wang P, Hao X, Li X, et al. Curcumin inhibits adverse psychological stress-induced proliferation and invasion of glioma cells via down-regulating the ERK/MAPK pathway. J Cell Mol Med 2021;25:7190-203. [Crossref] [PubMed]
- Tang F, Wang H, Chen E, et al. LncRNA-ATB promotes TGF-β-induced glioma cells invasion through NF-κB and P38/MAPK pathway. J Cell Physiol 2019;234:23302-14. [Crossref] [PubMed]
- Wang A, Meng M, Zhao X, et al. Long non-coding RNA ENST00462717 suppresses the proliferation, survival, and migration by inhibiting MDM2/MAPK pathway in glioma. Biochem Biophys Res Commun 2017;485:513-21. [Crossref] [PubMed]
- Gieryng A, Pszczolkowska D, Walentynowicz KA, et al. Immune microenvironment of gliomas. Lab Invest 2017;97:498-518. [Crossref] [PubMed]
- Fu W, Wang W, Li H, et al. Single-Cell Atlas Reveals Complexity of the Immunosuppressive Microenvironment of Initial and Recurrent Glioblastoma. Front Immunol 2020;11:835. [Crossref] [PubMed]
- Wei J, Chen P, Gupta P, et al. Immune biology of glioma-associated macrophages and microglia: functional and therapeutic implications. Neuro Oncol 2020;22:180-94. [PubMed]
- Chen Z, Hambardzumyan D. Immune Microenvironment in Glioblastoma Subtypes. Front Immunol 2018;9:1004. [Crossref] [PubMed]
- Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545:495-9. [Crossref] [PubMed]


