
Hypermethylated WASF2: tumor suppressive role in head and neck squamous cell carcinoma
Highlight box
Key findings
• We confirmed that enhanced WASF2 was associated with long overall survival time in HNSCC. Its level of promoter methylation had detrimental effects on patient survival. In addition, WASF2 was positively associated with tumor-killing immune cells, while WASF2 methylation was positively related to immunosuppressive cells and immune-inhibitors.
What is known and what is new?
• WASF2 was implicated in tumorigenesis and the development of several cancers, including pancreatic cancer breast cancer and lung adenocarcinoma. WASF2 was indeed required for T cell activation.
• The clinical value of WASF2 in HNSCC was analyzed comprehensively for the first time. We investigated further the dysregulated pathways and immune imbalance in hypermethylated WASF2 patients.
What is the implication, and what should change now?
• Our results were obtained by querying a database, which enables us to study gene function rapidly and provides a potential direction for biological experiments. Future experiments are required to confirm the role of WASF2 in tumor promotion and immunomodulatory effects.
Introduction
An estimated 600,000 new head and neck squamous cell carcinoma (HNSCC) cases are diagnosed annually worldwide, with a mortality rate of 40–50%. Recently, traditional risks such as excessive alcohol consumption and tobacco exposure have decreased, while the risk of human papilloma virus (HPV) infection has increased. HPV-positive oropharyngeal cancers, in particular, had a favorable clinical outcome (1). Therefore, it was essential for us to identify potential prognostic markers in HNSCC. HPV status should also be considered.
WASF2, also known as WAVE2 and SCAR2, is located on chromosome 1p36.11. The primary function of WASF family proteins was to regulate the actin cytoskeleton via the Arp2/3 complex and to drive membrane protrusion (2). In the WASF proteins family, WASF1 and WASF3 were restricted to brain tissues, whereas WASF2 was widely distributed in almost all tissues except for skeletal muscle cells (3). WASF2 was implicated in tumorigenesis and the development of several cancers, including pancreatic cancer (4), breast cancer (5) and lung adenocarcinoma (6). However, the clinical relevance of WASF2 in HNSCC has not yet been studied. WASF2 aberrant expression was mechanically mediated by microRNA, Rac and HSPC300 and others (7-9). In addition to the accumulation of genetic alterations, the epigenetic landscape also played a significant role in regulating the carcinogenesis of HNSCC (10). In addition, evasion of immune destruction was a crucial hallmark of cancer. Furthermore, immune checkpoint inhibitors have established a new milestone in the treatment of cancer (11,12). WASF2 was indeed required for T cell activation (13). Furthermore, DNA methylation inhibitors can boost the immunogenicity of tumor cells (14). Therefore, WASF2 DNA methylation may contribute to forming an immunosuppressive microenvironment.
In this study, the WASF2 level, WASF2 methylation level, and the relationship between WASF2 expression and methylation in HNSCC were analyzed comprehensively for the first time. We evaluated the prognostic value of WASF2, the methylation of the WASF2 promoter, and the association between WASF2 and clinicopathological features. We investigated further the dysregulated pathways and immune imbalance in hypermethylated WASF2 patients. We present the following article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1133/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DNMIVD database
DNMIVD (http://119.3.41.228/dnmivd/) is a visual DNA methylation database for 26 TCGA cancer types (15). The DNA methylation profile of various human cancer were compiled using high-throughput microarray data from the TCGA and GEO databases. Using the DNMIVD database, we investigated the WASF2 expression levels and WASF2 promoter methylation levels in tumor tissues and adjacent normal tissues, as well as the correlation between WASF2 promotor methylation levels and WASF2 transcriptional levels. In addition, the relationship between WASF2 promoter methylation and disease-free interval (DFI) and progression-free interval (PFI) was analyzed using median DNA methylation beta values as a cutoff to separate samples into high and low groups.
TIMER database
A user-friendly online database, TIMER (http://timer.cistrome.org/) provides dynamic analysis and visualization of immune infiltration levels via the TCGA database (16). WASF2 was plotted on Kaplan-Meier curves for HNSCC, HPV-positive HNSCC, and HPV-negative HNSCC. The threshold value for patients was 40%, and the follow-up time was 60 months. In addition, associations between WASF2 level and immune infiltrates in HNSCC, and HPV-positive HNSCC were investigated. In HNSCC and HPV-positive HNSCC, the correlation of WASF2 with specific immunocyte markers was also evaluated.
TNMplot database
The TNMplot database (https://tnmplot.com/analysis/) is an integrated online database to analyze differential gene expression in tumor, normal and metastatic tissues (17). Using this database, the expression level of WASF2 in tumor tissues and adjacent normal tissues was compared in human cancers.
GEO database
To determine whether DNA methylation regulated the expression of WASF2, we queried the GEO database. Expression profiling of malignant melanoma treated with the DNA methylation inhibitor 5-Aza-2'-deoxycytidine (5AzadC) (GEO accession number: GSE9118) was investigated. GEO2R was used to identify the change in WASF2 after treatment with a DNA methylation inhibitor in melanoma.
Kaplan-Meier Plotter database
Kaplan-Meier Plotter (http://kmplot.com/analysis/) is a website for analyzing survival analysis for 21 types of human cancer (18). The Kaplan-Meier Plotter database was used to analyze the association between WASF2 expression and survival in HNSCC, breast cancer, esophageal squamous cell carcinoma (ESCC), kidney renal clear cell carcinoma, bladder carcinoma, liver hepatocellular carcinoma, ovarian cancer, pancreatic ductal adenocarcinoma (PDAD), and kidney renal papillary cell. We determined that 60 months was the optimal cutoff for follow-up. The Kaplan-Meier Plotter database was also used to examine the relation between WASF2 and clinical characteristics.
MethSurv database
MethSurv database (https://biit.cs.ut.ee/methsurv/) is an interactive database for survival analysis of 25 different human cancers according to CpG methylation patter (19). Using the MethSurv database, survival analysis based on CpG sites of WASF2 in HNSCC was investigated.
Gene set enrichment analysis (GSEA)
According to median value of WASF2 expression levels, 522 HNSCC patients were divided into low and high-expression groups in the TCGA database. The pathways associated with the differential WASF2 expression were subsequently analyzed using GSEA.
Statistical analysis
Following the instructions, statistical analysis and graphic processing were conducted using GraphPad prism 8.0. In addition, two-sided, and P<0.05 were regarded as statistically significant difference.
Results
WASF2 expression negatively correlated with WASF2 methylation level in HNSCC
Using the DNMIVD database, WASF2 transcriptional levels were significantly down-regulated in HNSCC compared with the adjacent normal tissues (P<0.001, Figure 1A). We further confirmed that WASF2 expression was low in head and neck cancer using TNMplot database (P=0.018, Figure S1A) Nevertheless, patients with tongue cancer, a subtype of HNSCC, had elevated levels of WASF2 (P<0.001, Figure S1B). In addition, the expression levels of WASF2 were found to be higher in liver cancer (P<0.001, Figure S1C) and pancreas cancer (P<0.001, Figure S1D), while less in colon cancer (P<0.001, Figure S1E), breast cancer (P<0.001, Figure S1F), oesophageal cancer (P<0.001, Figure S1G), lung cancer (P<0.001, Figure S1H), and skin cancer (P=0.012, Figure S1I).
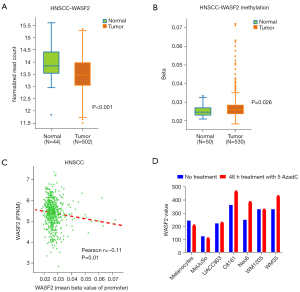
DNA methylation is aberrant in HNSCC and plays a crucial role in tumorigenesis (20). Using the DNMIVD database, we discovered that HNSCC patients had a higher level of WASF2 promoter methylation level than adjacent normal tissues (P=0.026, Figure 1B). Furthermore, WASF2 promoter methylation levels were negatively correlated with WASF2 expression, as predicted (Pearson r=−0.11, P=0.01, Figure 1C). To confirm the relationship between WASF2 methylation and WASF2 expression levels, we analyzed the expression change of WASF2 in melanoma cell lines treated with the DNA methylation inhibitor 5AzadC using the GEO database (GEO accession number: GSE9118). Results showed that four out of five malignant melanoma cell lines expressed more WASF2 after 48 hours of treatment with a methylation inhibitor (Figure 1D). In addition, WASF2 promoter methylation levels were associated negatively with esophageal carcinoma (Pearson r=−0.21, P=0.005, Figure S2A), breast invasive carcinoma (Pearson r=−0.16, P<0.001, Figure S2B), lung squamous cell carcinoma (Pearson r=−0.21, P<0.001, Figure S2C), kidney renal clear carcinoma (Pearson r=−0.15, P=0.006, Figure S2D), kidney renal papillary cell carcinoma (KIRP) (Pearson r=−0.27, P<0.001, Figure S2E) and skin cutaneous melanoma (Pearson r=−0.37, P<0.001, Figure S2F).
Prognostic value of WASF2 and WASF2 methylation in HNSCC
After investigating WASF2 transcriptional levels and WASF2 methylation levels in HNSCC, we investigated the prognostic value of WASF2 and WASF2 methylation in HNSCC. Through TIMER, the results revealed that HNSCC patients with higher levels of WASF2 had a longer overall survival (OS) time (HR=0.824, P=0.009, Figure 2A). We further investigated the prognosis of WASF2 in HPV-positive and HPV-negative HNSCC patients, given that HPV status is an important risk factor for developing HNSCC. We found that HPV-positive HNSCC patients with high-expressed WASF2 had a favorable OS time (HR=0.574, P=0.007, Figure 2B). Nevertheless, WASF2 was not a prognostic indicator in HPV-negative HNSCC patients (Figure 2C). Based on the PrognoScan database, we confirmed that enhanced WASF2 was associated with long OS time in HNSCC (Figure S3A). In addition, patients with higher levels of WASF2 had a longer OS time in breast cancer (Figure S3B), ESCC (Figure S3C), and KIRC (Figure S3D). However, patients with up-regulated WASF2 had a shorter OS in bladder carcinoma (BCA) (Figure S3E), liver hepatocellular carcinoma (Figure S3F), ovarian cancer (OV) (Figure S3G), and PDAD (Figure S3H). Moreover, up-regulated WASF2 was associated with a longer recurrence-free survival time (RFS) in KIRP (Figure S3I). In contrast, patients with elevated WASF2 had the worst RFS in liver hepatocellular carcinoma (Figure S3J), OV (Figure S1K), and PDAD (Figure S3L).
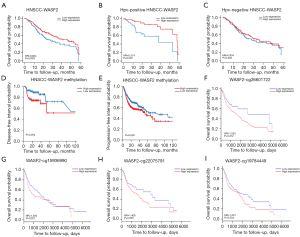
Using the DNMIVD database, the survival plot of WASF2 methylation in HNSCC patients was further investigated. The outcomes demonstrated that HNSCC patients with high WASF2 promoter methylation had a shorter DFI (P=0.054, Figure 2D) and PFI (P=0.026, Figure 2E). Methylation occurs predominantly in CpG-island regions. Then, using the MethSurv database, we conducted a survival analysis based on a single CpG methylation. The results revealed that HNSCC patients with high methylation of CpG cg26601722 (HR =1.525, P=0.012, Figure 2F), CpG cg15936990 (HR =1.394, P=0.027, Figure 2G), CpG cg22075791 (HR =1.408, P=0.033, Figure 2H), and CpG cg19784449 (HR =1.331, P=0.034, Figure 2I) had short OS values.
WASF2 expression in a stratified HNSCC population
Using the Kaplan-Meier Plotter database, we investigated the correlation between WASF2 expression and specific clinicopathological characteristics in HNSCC patients. For OS, WASF2 had a significant effect on HNSCC patients with the following clinicopathological characteristics: male (n=366, HR =0.59, 95% CI: 0.4–0.85, P=0.0049), clinical IV stage (n=259, HR =0.62, 95% CI: 0.42–0.9, P=0.012), grade 2 (n=298, HR =0.69, 95% CI: 0.48–0.99, P=0.041), low mutation burden (n=243, HR =0.57, 95% CI: 0.34–0.97, P=0.037) or high mutation burden (n=251, HR =0.65, 95% CI: 0.44–0.95, P=0.025). Furthermore, high–expressed WASF2 of clinical III stage (n=42, HR =0.22, 95% CI: 0.05–1.04, P=0.037) HNSCC patients had a longer RFS (Table 1). In addition, we evaluated the relationship between WASF2 and clinical characteristics in KIRC and breast cancer (Table S1). WASF2 had significant effects on OS in patients with KIRC and breast cancer, according to the findings. Higher WASF2 expression was associated with longer OS for female (n=186, HR =0.42, 95% CI: 0.24–0.72, P=0.001), male (n=344, HR =0.54, 95% CI: 0.36–0.81, P=0.003), clinical I stage (n=265, HR =0.44, 95% CI: 0.22–0.88, P=0.016), clinical IV stage (n=87, HR =0.58, 95% CI: 0.35–0.96, P=0.031), Grade 3 (n=206, HR =0.41, 95% CI: 0.25–0.69, P<0.001), or low mutation burden (n=168, HR =0.43, 95% CI: 0.24–0.79, P=0.005) KIRC patients. Moreover, up–regulated WASF2 brought about longer OS for ER positive (n=754, HR =0.61, 95% CI: 0.42–0.89, P=0.01), PR positive (n=156, HR =0.3, 95% CI: 0.09–1.02, P=0.041), HER2 negative (n=1,459, HR =0.58, 95% CI: 0.42–0.78, P<0.001) and luminal A subtype (n=794, HR =0.43, 95% CI: 0.27–0.69, P<0.001) or negative lymph node status (n=726, HR =0.45, 95% CI: 0.26–0.77, P=0.003) breast cancer patients.
Table 1
| Clinicopathological characteristic | OS (n=499) | RFS (n=124) | |||||
|---|---|---|---|---|---|---|---|
| N | Hazard ratio | P value | N | Hazard ratio | P value | ||
| Sex | |||||||
| Female | 133 | 1.6 (0.92–2.79) | 0.095 | 46 | 2.14 (0.58–7.86) | 0.24 | |
| Male | 366 | 0.59 (0.4–0.85) | 0.0049* | 78 | 0.18 (0.02–1.14) | 0.068 | |
| Stage | |||||||
| 1 | 25 | 2.57 (0.27–24.99) | 0.4 | 23 | 0.29 (0.04–2.46) | 0.23 | |
| 2 | 69 | 0.58 (0.25–1.34) | 0.2 | 43 | 3.93 (0.44–35.17) | 0.19 | |
| 3 | 78 | 1.8 (0.83–3.89) | 0.13 | 42 | 0.22 (0.05–1.04) | 0.037* | |
| 4 | 259 | 0.62 (0.42–0.9) | 0.012* | 0 | |||
| Grade | |||||||
| 1 | 61 | 0.35 (0.1–1.24) | 0.089 | 26 | 0.4 (0.06–2.46) | 0.3 | |
| 2 | 298 | 0.69 (0.48–0.99) | 0.041* | 69 | 0.36 (0.1–1.28) | 0.099 | |
| 3 | 119 | 0.58 (0.31–1.08) | 0.084 | 25 | 0 (0–lnf) | 0.076 | |
| Mutation burden | |||||||
| Low | 243 | 0.57 (0.34–0.97) | 0.037* | 73 | 0.47 (0.13–1.62) | 0.22 | |
| High | 251 | 0.65 (0.44–0.95) | 0.025* | 50 | 0.25 (0.03–2.01) | 0.16 | |
*, P<0.05. HNSCC, head and neck squamous cell carcinoma; OS, overall survival; RFS, recurrence-free survival.
Identification of WASF2-related signaling pathway via GSEA
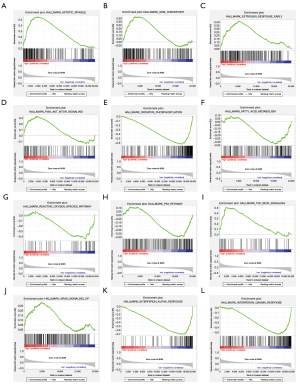
GSEA was used to analyze the biological function of WASF2 in HNSCC patients (Table 2). 24 out of 50 gene sets were upregulated in the high-expression WASF2 phenotype, and 11 gene sets were significantly enriched at NES >1.0, nominal P<0.05 and FDR q<0.25. In contrast, in the low-expression WASF2 phenotype, 26 out of 50 gene sets were upregulated, and 13 were significantly enriched at NES <1.0, nominal P<0.05 and FDR q<0.25. The significant hallmark gene sets involved in tumor initiation and development were the following: “MITOTIC SPINDLE” (Figure 3A), “G2M CHECKPOINT” (Figure 3B), “ESTROGEN RESPONSE EARLY” (Figure 3C), “PI3K AKT MTOR SIGNALING” (Figure 3D), “OXIDATIVE PHOSPHORYLATION” (Figure 3E), “FATTY ACID METABOLISM” (Figure 3F), “REACTIVE OXYGEN SPECIES PATHYWAY” (Figure 3G), and “P53 PATHWAY” (Figure 3H). The gene sets involved in immune response were the following: “TGF BETA SIGNALING” (Figure 3I), “KRAS SIGNALING” (Figure 3J), “INTERFERON ALPHA RESPONSE” (Figure 3K), and “INTERFERON GAMMA RESPONSE” (Figure 3L).
Table 2
| Gene set | Size | NES | NOM P value | FDR q value |
|---|---|---|---|---|
| Upregulated gene sets in high WASF2 expression group | ||||
| HALLMARK_MITOTIC_SPINDLE | 198 | 2.63 | 0 | 0 |
| HALLMARK_UV_RESPONSE_DN | 141 | 2.2 | 0 | 0 |
| HALLMARK_TGF_BETA_SIGNALING | 54 | 1.99 | 0.002 | 0 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 197 | 1.96 | 0 | 0 |
| HALLMARK_PROTEIN_SECRETION | 95 | 1.83 | 0 | 0.001 |
| HALLMARK_ANDROGEN_RESPONSE | 99 | 1.78 | 0 | 0.002 |
| HALLMARK_G2M_CHECKPOINT | 195 | 1.75 | 0 | 0.002 |
| HALLMARK_HEDGEHOG_SIGNALING | 36 | 1.55 | 0.028 | 0.013 |
| HALLMARK_HEME_METABOLISM | 194 | 1.46 | 0.003 | 0.029 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 105 | 1.41 | 0.007 | 0.039 |
| HALLMARK_KRAS_SIGNALING_UP | 199 | 1.35 | 0.013 | 0.066 |
| Upregulated gene sets in low WASF2 expression group | ||||
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 200 | −2.89 | 0 | 0 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 95 | −2.86 | 0 | 0 |
| HALLMARK_MYC_TARGETS_V1 | 196 | −2.48 | 0 | 0 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 198 | −2.46 | 0 | 0 |
| HALLMARK_MYC_TARGETS_V2 | 58 | −2.44 | 0 | 0 |
| HALLMARK_DNA_REPAIR | 149 | −2.27 | 0 | 0 |
| HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY | 49 | −1.61 | 0.005 | 0.009 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 110 | −1.59 | 0.007 | 0.008 |
| HALLMARK_ADIPOGENESIS | 198 | −1.54 | 0.002 | 0.014 |
| HALLMARK_MYOGENESIS | 199 | −1.53 | 0 | 0.014 |
| HALLMARK_UV_RESPONSE_UP | 156 | −1.47 | 0.01 | 0.025 |
| HALLMARK_FATTY_ACID_METABOLISM | 157 | −1.46 | 0.003 | 0.025 |
| HALLMARK_P53_PATHWAY | 196 | −1.32 | 0.03 | 0.085 |
GSEA, Gene Set Enrichment Analysis; HNSCC, head and neck squamous cell carcinoma; NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate.
WASF2 and WASF2 methylation disrupted immune balance in HNSCC
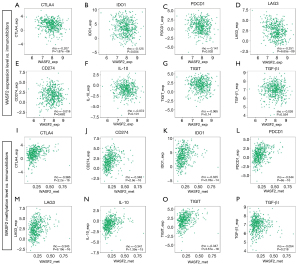
According to the results of the GSEA, WASF2 is an immune-related gene. Given that WASF2 was a favorable predicator in HNSCC, particularly in HPV-positive HNSCC patients, but not in HPV-negative HNSCC patients. We evaluated the correlation between WASF2 and specific infiltrating immune cells using TIMER database both in HNSCC and HPV-positive HNSCC. WASF2 was positively correlated with B cells (r=0.213, P<0.001, Figure 4A), CD4+ T cells (r=0.262, P<0.001, Figure 4B) and dendritic cells (r=0.141, P<0.001, Figure 4C), but not with CD8+ T cells (Figure 4D), macrophages (Figure 4E) and neutrophils (Figure 4F) among HNSCC patients. Meanwhile, WASF2 was positively correlated with B cells (r=0.284, P=0.011, Figure 4G), CD8+ T cells (r=0.227, P=0.046, Figure 4H), CD4+ T cells (r=0.336, P<0.001, Figure 4I) and dendritic cells (r=0.276, P=0.011, Figure 4J), but not with macrophages (Figure 4K) and neutrophils (Figure 4L) among patients with HPV-positive HNSCC. In addition, the association between WASF2 and additional immune cell markers both in HNSCC and HPV-positive HNSCC was evaluated. In HNSCC, we discovered a significant correlation between WASF2 and markers of TAM, M1 Macrophages, Tfh, Th17 and Treg cells. In HPV-positive HNSCC, WASF2 was also correlated with markers of M1 macrophages, Tfh and Th17 cells (Table 3). The correlation between WASF2 and tumor-killing immune cells was generally positive, indicating that WASF2 may promote the anti-tumor immune response. In contrast, WASF2 methylation was positively correlated with immature B cells (Figure 4M), Th17 (Figure 4N), Treg (Figure 4O), myeloid derived suppressor cells (Figure 4P), and Th2 (Figure 4Q), which may modulate immunosuppressive activity.

Table 3
| Description | Gene markers | HNSCC | HPV + HNSCC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | |||||||||
| Cor | P | Cor | P | Cor | P | Cor | P | |||||
| CD8+ T cell | CD8A | 0.012 | 0.177 | 0.07 | 0.122 | 0.212 | * | 0.273 | ** | |||
| CD8B | −0.007 | 0.874 | 0.033 | 0.459 | 0.191 | 0.0592 | 0.218 | * | ||||
| T cell (general) | CD3D | −0.027 | 0.54 | 0.021 | 0.646 | 0.151 | 0.137 | 0.21 | 0.483 | |||
| CD3E | 0.09 | * | 0.154 | *** | 0.256 | * | 0.341 | |||||
| CD2 | 0.058 | 0.186 | 0.112 | * | 0.222 | * | 0.298 | ** | ||||
| B cell | CD19 | 0.146 | *** | 0.198 | *** | 0.264 | ** | 0.361 | *** | |||
| CD79A | 0.215 | *** | 0.271 | *** | 0.304 | ** | 0.422 | *** | ||||
| Monocyte | CD86 | 0.068 | 0.12 | 0.129 | ** | 0.018 | 0.86 | 0.141 | 0.188 | |||
| CSF1R | 0.105 | * | 0.173 | *** | 0.09 | 0.377 | 0.191 | 0.073 | ||||
| TAM | CCL2 | 0.098 | * | 0.136 | ** | 0.052 | 0.613 | 0.078 | 0.465 | |||
| CD68 | 0.181 | *** | 0.229 | *** | −0.001 | 0.992 | 0.042 | 0.697 | ||||
| IL-10 | 0.098 | * | 0.171 | *** | 0.066 | 0.52 | 0.144 | 0.178 | ||||
| M1 Macrophage | NOS2 | 0.346 | *** | 0.325 | *** | 0.349 | *** | 0.358 | *** | |||
| IRF5 | 0.235 | *** | 0.231 | *** | 0.258 | * | 0.244 | * | ||||
| PTGS2 | 0.181 | *** | 0.168 | *** | 0.326 | ** | 0.332 | ** | ||||
| M2 Macrophage | CD163 | 0.033 | 0.454 | 0.096 | * | −0.124 | 0.225 | −0.077 | 0.475 | |||
| VSIG4 | −0.005 | 0.913 | 0.044 | 0.335 | −0.104 | 0.308 | −0.08 | 0.454 | ||||
| MS4A4A | −0.023 | 0.6 | 0.031 | 0.499 | −0.13 | 0.201 | −0.088 | 0.41 | ||||
| Natural killer cell | KIR2DL1 | 0.034 | 0.434 | 0.068 | 0.133 | 0.026 | 0.797 | 0.093 | 0.388 | |||
| KIR2DL3 | 0.002 | 0.969 | 0.015 | 0.741 | 0.097 | 0.343 | 0.1 | 0.352 | ||||
| KIR2DL4 | 0.002 | 0.965 | 0.056 | 0.212 | 0.06 | 0.558 | 0.159 | 0.137 | ||||
| KIR3DL1 | 0.119 | ** | 0.153 | *** | 0.077 | 0.451 | 0.145 | 0.174 | ||||
| KIR3DL2 | 0.09 | * | 0.118 | ** | 0.175 | 0.0843 | 0.236 | * | ||||
| KIR3DL3 | 0.057 | 0.194 | 0.057 | 0.206 | 0.091 | 0.374 | 0.117 | 0.275 | ||||
| KIR2DS4 | 0.006 | 0.894 | 0.051 | 0.261 | 0.087 | 0.392 | 0.183 | 0.0853 | ||||
| Dendritic cell | HLA-DPB1 | 0.006 | 0.889 | 0.061 | 0.176 | 0.076 | 0.457 | 0.154 | 0.15 | |||
| HLA-DQB1 | −0.004 | 0.933 | 0.027 | 0.55 | 0.132 | 0.195 | 0.216 | * | ||||
| HLA-DRA | 0.036 | 0.417 | 0.094 | * | 0.115 | 0.26 | 0.212 | * | ||||
| HLA-DPA1 | 0.045 | 0.305 | 0.097 | * | 0.124 | 0.225 | 0.219 | * | ||||
| CD1C | 0.218 | *** | 0.28 | *** | 0.316 | ** | 0.377 | *** | ||||
| NRP1 | 0.138 | ** | 0.189 | *** | −0.012 | 0.904 | −7 | 0.948 | ||||
| ITGAX | 0.114 | ** | 0.184 | *** | 0.053 | 0.603 | 0.174 | 0.103 | ||||
| Th1 | TBX21 | 0.05 | 0.254 | 0.098 | * | 0.19 | 0.0615 | 0.256 | * | |||
| STAT4 | 0.059 | 0.178 | 0.108 | * | 0.145 | 0.153 | 0.243 | * | ||||
| STAT1 | −0.007 | 0.868 | 0.042 | 0.349 | 0.054 | 0.598 | 0.11 | 0.303 | ||||
| IFNG | −0.084 | 0.0548 | −0.043 | 0.345 | 0.097 | 0.343 | 0.135 | 0.207 | ||||
| TNF | 0.14 | ** | 0.16 | *** | 0.252 | * | 0.308 | ** | ||||
| Th2 | GATA3 | 0.021 | 0.633 | 0.071 | 0.116 | 0.172 | 0.0896 | 0.245 | * | |||
| STAT6 | 0.45 | *** | 0.444 | *** | 0.434 | *** | 0.435 | *** | ||||
| STAT5A | 0.268 | *** | 0.287 | *** | 0.167 | 0.101 | 0.243 | * | ||||
| IL13 | −0.024 | 0.582 | −0.029 | 0.526 | 0.043 | 0.675 | 0.123 | 0.249 | ||||
| Tfh | BCL6 | 0.501 | *** | 0.477 | *** | 0.455 | *** | 0.462 | *** | |||
| Th17 | STAT3 | 0.511 | *** | 0.526 | *** | 0.556 | *** | 0.566 | *** | |||
| IL17A | 0.163 | *** | 0.189 | *** | 0.349 | *** | 0.369 | *** | ||||
| Treg | FOXP3 | 0.241 | *** | 0.809 | *** | 0.333 | *** | 0.441 | *** | |||
| CCR8 | 0.323 | *** | 0.387 | *** | 0.457 | *** | 0.541 | *** | ||||
| STAT5B | 0.323 | *** | 0.34 | *** | 0.154 | 0.13 | 0.201 | 0.0589 | ||||
| TGFB1 | 0.094 | * | 0.153 | *** | −0.1 | 0.327 | −0.074 | 0.488 | ||||
*, P<0.05; **, P<0.01; ***, P<0.001. HNSCC, head and neck squamous cell carcinoma; HPV, human papilloma virus; TAM, tumor-associated macrophage; Th, T helper cell; Tfh, Follicular helper T cell; Treg, regulatory T cell; Cor, R value of Spearman’s correlation; None, correlation without adjustment. Purity, correlation adjusted by purity.
Correlation of WASF2 transcriptional expression/methylation levels with immune-inhibitors in HNSCC
There is widespread agreement that HNSCC is a highly immunosuppressive disease (21). Using the TISIDB database, the association between WASF2 expression/methylation levels and immunosuppressive markers was evaluated. WASF2 expression levels correlated negatively with CTLA-4 (r=−0.207, P<0.001, Figure 5A), IDO1 (r=−0.125, P=0.004, Figure 5B), PDCD1 (r=−0.141, P=0.002, Figure 5C) and LAG3 (r=−0.251, P<0.001, Figure 5D), but not with CD274 (Figure 5E), IL-10 (Figure 5F), TIGIT (Figure 5G), and TGF-β1 (Figure 5H). Expertly, WASF2 methylation levels were positively correlated with CTLA4 (r=−0.366, P<0.001, Figure 5I), CD274 (r=−0.348, P<0.001, Figure 5J), IDO1 (r=−0.325, P<0.001, Figure 5K), PDCD1 (r=−0.344, P<0.001, Figure 5L), LAG3 (r=−0.343, P<0.001, Figure 5M), IL-10 (r=−0.341, P<0.001, Figure 5N), and TIGIT (r=−0.347, P<0.001, Figure 5O), but not with TGF-β1 (Figure 5P). It suggested that targeting hypermethylated WASF2 facilitated immune checkpoint-based therapies.
Discussion
WASF2’s combination of expression and methylation has yet to be clarified. This study analyzed WASF2 expression and WASF2 methylation in HNSCC using online databases and bioinformatics data mining tools. WASF2 transcriptional levels were low in HNSCC and negatively correlated with WASF2 promoter methylation, according to our findings. According to the GEO database, the DNA methylation inhibitor 5AzadC increased transcriptional levels of WASF2. Patients with HNSCC and high WASF2 expression had a prolonged OS. Patients whose WASF2 promoters were more heavily methylated had a poor prognosis. Interestingly, HPV-positive HNSCC patients with high WASF2 expression had favorable prognosis compared with HPV-negative HNSCC patients. High-expressed WASF2 of HNSCC patients characterized by male, clinical stage IV, grade 2, and mutation burden had a longer OS. High levels of expressed WASF2 were associated with a longer RFS in HNSCC patients with clinical III stage. WASF2 was implicated in tumor procession and immune response, according to the results of GSEA. Thus, we examined the relationship between WASF2 and WASF2 methylation and the tumor immune microenvironment. WASF2 was generally positively correlated with tumor-killing immune cells, whereas WASF2 methylation was consistently positively associated with immunosuppressive cells. Strikingly, WASF2 had negative correlation with specific immunoinhibitory (CTLA-4, IDO1, PDCD1 and LAG3) while WASF2 methylation had positive correlation with specific immunoinhibitory (CTLA-4, CD274, IDO1, PDCD1, LAG3, IL-10 and TIGIT) in HNSCC. These findings suggested that hypermethylated WASF2 was a potential tumor-suppressor gene and that hypermethylated WASF2 could suppress tumor progression in HNSCC by regulating tumor growth and immune microenvironment balance.
WASF2 is required for tumor cell invasion and metastasis by regulating the actin cytoskeleton (8). WASF2 is widely expressed in gastric cancer (22), hepatocellular carcinoma (23) and pancreatic cancer (24). WASF2 consistently increased in liver cancer and pancreatic cancer while decreasing in HNSCC. In addition, miR-146a could inhibit WASF2 expression in gastric cancer, and WASF2 protein levels were negatively correlated with miR-146a levels (7). Similarly, we found WASF2 expression was negatively associated with WASF2 methylation levels in HNSCC. Owing to 5AzadC incorporation into DNA and inhibition of DNA methylation (25), it was confirmed that WASF2 methylation suppresses WASF2 expression.
The study found that PDAC patients with high-expressed WASF2 had significantly shorter OS times (26). Moreover, WASF2 was a poor prognostic indicator of hepatocellular carcinoma (23). In LIHC and PDAC, up-regulated WASF2 was consistently associated with a short OS and RFS. Patients with HNSCC who had more WASF2 had a longer OS time. Several gene-specific DNA methylation have been identified for diagnosis and prognosis in HNSCC (27). Herein, we found that patients with a hypermethylated WASF2 promoter had poor PFI. In HNSCC, we also identified four prognosis-associated CpG-islands of WASF2.
Mechanically, WASF2 was crucial for actin polymerization, which is essential for cell migration. WASF2-deficient mice could decrease sprouting and branching of endothelial cells during angiogenesis (28). In addition, WASF2 works in controlling epidermal shape and growth via Wnt/β-catenin signaling (29). A study discovered that WASF2 contributed to breast epithelial morphology and that knocking down WASF2 promoted the proliferation of late-stage acini and characterized a partial EMT-like phenotype (30). Similarly, we discovered that WASF2 was associated with cell proliferation-related pathways in HNSCC, including mitotic spindle assembly, G2/M checkpoints, and p53 pathway.
There existed broad differences in the tumor-infiltrating immune population of HPV-positive and HPV-negative HNSCC. In HPV-positive HNSCC, higher frequencies of infiltrating B cells and helper CD4+ T cells were observed (31). We discovered that HPV-positive HNSCC patients had a stronger correlation with B cells and CD4+ T cells than HNSCC patients. In addition, WASF2 was a critical component for T cell activation because it regulated actin cytoskeletal reorganization, intracellular calcium signaling, and NFκB activation (32). We discovered an intriguing correlation between WASF2 methylation levels, specific immunosuppressive cells, and immune checkpoints. In a mouse model of ovarian cancer, treatment with anti-PD-L1 and 5-AZAdC could increase infiltrating CD8+ T cells and Th1-type chemokine expression (33). In addition, WASF2 can maintain T cell homeostasis by inhibiting mTOR activation. A conditional WASF2 knockout in T cells could impair antigen-specific T cell response and result in severe autoimmunity (34). According to our findings, patients with hypermethylated WASF2 disrupted the intratumoral immune homeostasis in HNSCC and HPV-positive HNSCC. Our results were obtained by querying a database, which enables us to study gene function rapidly and efficiently and provides a potential direction for biological experiments. Future experiments are required to confirm the role of WASF2 in tumor promotion and immunomodulatory effects.
Conclusions
We determined the potential clinical relevance of WASF2 hypermethylation in HNSCC. GSEA results indicated that WASF2 was involved in tumor growth and the immune response. The correlation between WASF2 methylation and tumor microenvironment also sheds light on WASF2’s role as an immunomodulator in innate and adaptive immune responses. By accumulating its genetic alterations, methylation abnormalities, and immune microenvironment imbalance, we could demonstrate that WASF2 played a significant role in HNSCC.
Acknowledgments
We thank the uploaders of Bilibili website for selflessly sharing the bioinformation methods.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1133/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-1133/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269-82. [Crossref] [PubMed]
- Machesky LM, Mullins RD, Higgs HN, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A 1999;96:3739-44. [Crossref] [PubMed]
- Suetsugu S, Miki H, Takenawa T. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun 1999;260:296-302. [Crossref] [PubMed]
- Taniuchi K, Furihata M, Naganuma S, et al. WAVE2 is associated with poor prognosis in pancreatic cancers and promotes cell motility and invasiveness via binding to ACTN4. Cancer Med 2018;7:5733-51. [Crossref] [PubMed]
- Yokotsuka M, Iwaya K, Saito T, et al. Overexpression of HER2 signaling to WAVE2-Arp2/3 complex activates MMP-independent migration in breast cancer. Breast Cancer Res Treat 2011;126:311-8. [Crossref] [PubMed]
- Semba S, Iwaya K, Matsubayashi J, et al. Coexpression of actin-related protein 2 and Wiskott-Aldrich syndrome family verproline-homologous protein 2 in adenocarcinoma of the lung. Clin Cancer Res 2006;12:2449-54. [Crossref] [PubMed]
- Yao Q, Cao Z, Tu C, et al. MicroRNA-146a acts as a metastasis suppressor in gastric cancer by targeting WASF2. Cancer Lett 2013;335:219-24. [Crossref] [PubMed]
- Kurisu S, Suetsugu S, Yamazaki D, et al. Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene 2005;24:1309-19. [Crossref] [PubMed]
- Cai X, Xiao T, James SY, et al. Metastatic potential of lung squamous cell carcinoma associated with HSPC300 through its interaction with WAVE2. Lung Cancer 2009;65:299-305. [Crossref] [PubMed]
- Johnson DE, Burtness B, Leemans CR, et al. Head and neck squamous cell carcinoma. Nat Rev Dis Primers 2020;6:92. [Crossref] [PubMed]
- de Miguel M, Calvo E. Clinical Challenges of Immune Checkpoint Inhibitors. Cancer Cell 2020;38:326-33. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Joseph N, Biber G, Fried S, et al. A conformational change within the WAVE2 complex regulates its degradation following cellular activation. Sci Rep 2017;7:44863. [Crossref] [PubMed]
- Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015;162:974-86. [Crossref] [PubMed]
- Ding W, Chen J, Feng G, et al. DNMIVD: DNA methylation interactive visualization database. Nucleic Acids Res 2020;48:D856-62. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Bartha Á, Győrffy B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int J Mol Sci 2021; [Crossref] [PubMed]
- Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11:6047. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Pan Y, Song Y, Cheng L, et al. Analysis of methylation-driven genes for predicting the prognosis of patients with head and neck squamous cell carcinoma. J Cell Biochem 2019;120:19482-95. [Crossref] [PubMed]
- Gavrielatou N, Doumas S, Economopoulou P, et al. Biomarkers for immunotherapy response in head and neck cancer. Cancer Treat Rev 2020;84:101977. [Crossref] [PubMed]
- Yao Q, Tu C, Lu D, et al. Clinicopathological significance of the microRNA-146a/WASP-family verprolin-homologous protein-2 axis in gastric cancer. Cancer Sci 2017;108:1285-92. [Crossref] [PubMed]
- Yang LY, Tao YM, Ou DP, et al. Increased expression of Wiskott-Aldrich syndrome protein family verprolin-homologous protein 2 correlated with poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2006;12:5673-9. [Crossref] [PubMed]
- Kitagawa T, Taniuchi K, Tsuboi M, et al. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol 2019;13:212-27. [Crossref] [PubMed]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002;21:5483-95. [Crossref] [PubMed]
- Yamazaki D, Suetsugu S, Miki H, et al. WAVE2 is required for directed cell migration and cardiovascular development. Nature 2003;424:452-6. [Crossref] [PubMed]
- Martone T, Gillio-Tos A, De Marco L, et al. Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res 2007;13:5089-94. [Crossref] [PubMed]
- Bryce NS, Reynolds AB, Koleske AJ, et al. WAVE2 regulates epithelial morphology and cadherin isoform switching through regulation of Twist and Abl. PLoS One 2013;8:e64533. [Crossref] [PubMed]
- Cohen J, Raviv S, Adir O, et al. The Wave complex controls epidermal morphogenesis and proliferation by suppressing Wnt-Sox9 signaling. J Cell Biol 2019;218:1390-406. [Crossref] [PubMed]
- Jia S, Jia Y, Weeks HP, et al. Down-regulation of WAVE2, WASP family verprolin-homologous protein 2, in gastric cancer indicates lymph node metastasis and cell migration. Anticancer Res 2014;34:2185-94. [PubMed]
- Cillo AR, Kürten CHL, Tabib T, et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020;52:183-199.e9. [Crossref] [PubMed]
- Nolz JC, Gomez TS, Zhu P, et al. The WAVE2 complex regulates actin cytoskeletal reorganization and CRAC-mediated calcium entry during T cell activation. Curr Biol 2006;16:24-34. [Crossref] [PubMed]
- Peng D, Kryczek I, Nagarsheth N, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015;527:249-53. [Crossref] [PubMed]
- Liu M, Zhang J, Pinder BD, et al. WAVE2 suppresses mTOR activation to maintain T cell homeostasis and prevent autoimmunity. Science 2021;371:eaaz4544. [Crossref] [PubMed]


