
BTN2A1-BRAF fusion may be a novel mechanism of resistance to osimertinib in lung adenocarcinoma: a case report
Highlight box
Key findings
• BTN2A1-BRAF fusion serves as a mechanism of acquired resistance to osimertinib in NSCLC.
What is known and what is new?
• Osimertinib is a third-generation EGFR-TKI that targets EGFR sensitive mutation and exon20 T790M in NSCLC. Although some mechanisms of resistance to osimertinib have been documented, many remain unexplored, warranting further research.
• BTN2A1-BRAF fusion is a new mechanism of acquired resistance to osimertinib. Patients with EGFR sensitive mutation and BTN2A1-BRAF fusion derive little benefit from PD-L1 inhibitors.
What is the implication, and what should change now?
• Next-generation sequencing is indicated for NSCLC patients before treatment with EGFR-TKI to identify gene mutations when the disease status changes. It is highly recommended to adjust the treatment regimen for patients positive for BTN2A1-BRAF fusion when taking osimertinib.
Introduction
Lung cancer is the main cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for a significant portion of lung cancer cases (1). Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) have attracted significant interest in recent years, given their ability to effectively prevent disease progression. Osimertinib is a third-generation EGFR-TKI that can target exon20 T790M mutation. However, acquired resistance (AR) inevitably appears after 9–13 months of treatment. Although certain resistance mechanisms of third-generation EGFR-TKIs have been identified (such as C797S and MET amplification), many factors remain unknown, warranting exploration. Somatic mutations in BRAF genes occur in 3–8% of lung adenocarcinomas (2), and BRAF fusions account for 0.2% of NSCLCs (3). In our case, a new BRAF fusion, the butyrophilin subfamily 2 member A1 (BTN2A1)-BRAF fusion, was detected using next-generation sequencing (NGS) after resistance to osimertinib, and the patient was switched to programmed cell death-ligand 1 (PD-L1) inhibitors (duvalizumab). We present the following case in accordance with the CARE reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2060/rc).
Case presentation
A 53-year-old female who had never smoked was admitted to our hospital with a 2-month history of left-chest pain which worsened for 1 week. A computed tomography (CT) scan revealed space-occupying lesions in the left pulmonary hilum. Magnetic resonance imaging (MRI) showed a nodular abnormal signal intensity in the left frontal lobe, which was suspected to be a metastatic lesion. A biopsy sample obtained in the left-lobe of her lung by endobronchial ultrasonography (EBUS) was made into formalin-fixed paraffin-embedding (FFPE) tissue. The pathological diagnosis was NSCLC. Immunohistochemistry (IHC) staining was positive for TTF-1 but negative for P40 (Figure 1A). Finally, the patient was diagnosed with stage IIIc (T3N3Mx) lung adenocarcinoma. EGFR exon 19 deletion was detected in the tumor biopsy sample by 8-gene panel NGS (Burning Rock Dx, China). The patient first received monotherapy with gefitinib, and which yielded a good initial response. Space-occupying lesions in the left upper lobe of the lung regressed, and the area of inflammation in the surrounding pulmonary tissue decreased. MRI showed that the metastatic lesion in the left frontal lobe regressed. After 13 months, disease progression was observed with an enlarged irregular mass in the left lung lobe, several obstructive inflammation pneumonia, involvement of multiple lymph nodes, and left pleural effusion. A plasma circulating tumor DNA (ctDNA)-associated 9-gene panel (Anoroad, China) and droplet digital polymerase chain reaction (ddPCR) were performed to determine subsequent therapy. TP53 p.K132R and EGFR exon20 T790M-negative were identified in addition to the previously identified EGFR exon 19 deletion by liquid biopsy analysis. Despite treatment with “bevacizumab plus carboplatin plus pemetrexed” for 4 cycles, disease progression was observed. The left upper lobe mass became larger and obstructive pneumonia was exacerbated. Small nodules in the right lung and left oblique lung fissure were observed. Some lymph nodes under the left clavicle, in the mediastinum and at the left hilum of the lung were enlarged and were suspected to be metastatic. Moreover, left pleural effusion was still present. Besides, the patient experienced toxic side effects of chemotherapy with an abnormally elevated γ-Glutamyl transpeptidase (76 U/L), treated with the hepatoprotective drug glutathione. Besides, the patient experience other adverse events, including vomiting, allergy and urinary retention. EGFR amplification, MTOR mutation, and BRCA mutation were identified in the tumor biopsy sample using a 56-gene panel (Burning Rock Dx), the sample was harvested from the tumor tissue in the left lung by pulmonary puncture guided by CT. Later, the patient received a single cycle of “bevacizumab plus sintilimab”. Simultaneously, the EGFR exon20 T790M mutation was identified by cell-free DNA (cfDNA) using a 9-gene panel (Anoroad, China). Based on the liquid biopsy results, bevacizumab and osimertinib were administrated. The patient responded well and was diagnosed with stable disease (SD). Follow-up CT scans demonstrated regression of the primary lung lesion, resolution of the pleural effusion; and atelectasis recovery to a certain extent. CT scans also showed regression of the lymph nodes and left pleural thickening. The oncologist decided to continue treatment with osimertinib. At seven months, disease progression was follow-up, with enlargement of the primary lung tumor. Significant left pleural effusion was observed, accompanied by pericardial effusion. Part of the pleural effusion was encapsulated, with atelectasis of the left lung adjacent to the pleural effusion. Interestingly, we identified BTN2A1-BRAF fusion, a low tumor mutational burden (TMB; 3.84 Muts/Mb), and microsatellite stability (MSS), TGFBR1 p.D104Y, MYC amplification and FANCG amplification in the following 1021 cancer-related genes (Geneplus, China) using FFPE tumor tissues obtained by EBUS. A high PD-L1 expression (tumor proportion score, TPS =90%) was found by IHC. Therefore, the patient was given duvalizumab; however, she passed away three months later. Figure 1B depicts the entire course of treatment. Figure 2A,2B illustrate the model of BTN2A1-BRAF fusion. Table 1 shows the results of all tests conducted.
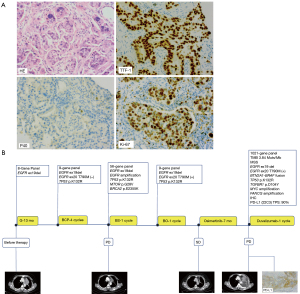
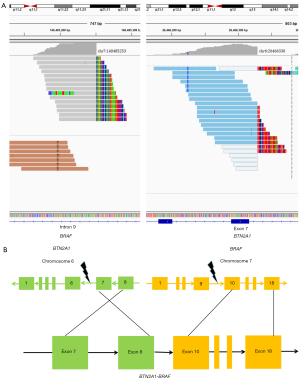
Table 1
| Time of sampling | Method | Sample | Gene | Mutation style | Frequency or CN |
|---|---|---|---|---|---|
| Before using EGFR-TKI | 8-gene panel NGS | FFPE | EGFR | Exon 19 deletion | 14.85% |
| After using gefitinib | ddPCR | cfDNA (liquid biopsy) | EGFR | Exon 20 T790M (−) | – |
| 9-gene panel NGS | cfDNA (liquid biopsy) | EGFR | Exon 19 deletion | 1.6% | |
| TP53 | p.K132R | 0.64% | |||
| After using bevacizumab + carboplatin + pemetrexed | 56-gene panel NGS | FFPE | EGFR | Exon 19 deletion | 26.02% |
| TP53 | p.K132R | 51.12% | |||
| EGFR | Amplification | CN =15.48 | |||
| mTOR | p.G28V | 62.79% | |||
| BRCA2 | p.E2355K | 70.17% | |||
| After using bevacizumab + sindilimab | 9-gene panel NGS | cfDNA (liquid biopsy) | EGFR | Exon 19 deletion | 8.2% |
| Exon 20 T790M (+) | 11.6% | ||||
| TP53 | p.K132R | 1.7% | |||
| After using osimertinib | 1021-gene panel NGS | FFPE+ blood | TMB | – | 3.84 Muts/Mb |
| MSI | MSS | – | |||
| EGFR | Exon19 deletion | 30.8% | |||
| Exon 20 T790M (+) | 14.1% | ||||
| BRAF | BTN2A1-BRAF fusion | 9.1% | |||
| TP53 | p.K132R | 30% | |||
| TGFBR1 | p.D104Y | 12.8% | |||
| MYC | Amplification | CN =7.4 | |||
| FANCG | Amplification | CN =6.8 | |||
| IHC | FFPE | PD-L1 | PD-L1 (+) | 90% |
CN, copy number; EGFR-TKI, epidermal growth factor receptor tyrosine kinase inhibitor; NGS, next generation sequencing; FFPE, formaldehyde fixed paraffin embedded; ddPCR, droplet digital polymerase chain reaction; cfDNA, cell free DNA; TMB, tumor mutational burden; MSI, microsatellite instability; MSS, microsatellite stability; IHC, immunohistochemistry; PD-L1, programmed cell death-ligand 1.
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the relatives of the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
To our knowledge, this is the first report of BTN2A1 being a fusion partner for BRAF and the clinical course of an NSCLC patient with BTN2A1-BRAF fusion. During therapy, BTN2A1-BRAF fusion was detected only after disease progression with osimertinib; therefore, we hypothesize that BTN2A1-BRAF fusion is another possible mechanism of AR to osimertinib. After EGFR T790M mutation, the following mutations were observed, including BTN2A1-BRAF fusion, TP53 p.K132R, TGFBR1 p.D104y, MYC amplification and FANCG amplification in addition to EGFR exon 19 deletion in the final 1021 panel by NGS. According to Le et al. (4) resistance mechanisms to osimertinib in patients with EGFR T790M include EGFR reactivation or bypass pathway. In our case, the acquisition of BTN2A1-BRAF fusion was observed. We hypothesize that BTN2A1-BRAF fusion may be the mechanism that leads to resistance to osimertinib. Schoenfeld et al. (5) found that TRIM24-BRAF fusion was acquired after disease progression following treatment with first-line osimertinib in NSCLC, while genomic alterations associated with BRAF fusion after later-line osimertinib were AGK-BRAF and MRPS33-BRAF. Both AGK-BRAF and MRPS33-BRAF were observed in patients with EGFR T790M loss. Inconsistent with the literature, in our case, BTN2A1-BRAF fusion was acquired in a patient who still harbored the EGFR T790M mutation after disease progression with osimertinib.
BRAF fusion genes differ in the position of the breakpoints within BRAF, which occur in introns (6). In this case, the breakpoint of BTN2A1-BRAF fusion occurred at intron 6 of the BTN2A1 gene and intron 9 of the BRAF gene (Figure 2). This structure led to oncogene overexpression due to the absence of the auto-inhibition domain in conserved region 1 (CR1) (7), located on the 5’ gene of BRAF. A promoter regulates oncogene expression in the 5’ gene derived from the fusion partner (8). Many experiments indicated that BRAF fusion could interfere with the RAS-RAF-MAPK pathway and compromise the efficacy of osimertinib (9,10). Vojnic et al. validated that AGK-BRAF and PJA2-BRAF fusions were mechanisms of AR to osimertinib and demonstrated that the combination of osimertinib and trametinib (MEK inhibitor) effectively inhibited the growth of BRAF-fusion-positive cell lines (11). Wang et al. reported a successful case of trametinib response in a patient with primary LIMD1-BRAF fusion, with progression-free survival (PFS) of 7.4 months (12). Overall, BTN2A1-BRAF fusion may be an underlying resistance mechanism to osimertinib.
In our case, the patient harbored exon 19 deletion, a sensitive EGFR mutation in NSCLC. Peng et al. argued that BRAF fusion was more likely to occur in patients with EGFR exon 19 deletion than those with non-exon 19 deletion mutations, such as exon 21 L858R and EGFR amplification (13). Schrock et al. reported ten NSCLC cases with BRAF fusion, including six with exon 21 L858R and four with exon 19 deletion (14). However, it remains unclear which baseline sensitizing mutation (exon 19 deletion or exon 21 L858R) is more frequently retained with BRAF fusion.
In this case, the patient first took gefitinib for 13 months. Despite a good initial response, disease progression was eventually observed, which prompted switching to osimertinib. However, growing evidence suggests that osimertinib yields a longer PFS and overall survival (OS) (15), and better response for patients with central nervous system (CNS) metastases than gefitinib (16). However, there was no significant difference in PFS and OS among Asian patients (16), and the beneficial effect of osimertinib in this patient population is not sustained.
Interestingly, in some cases, immune checkpoint inhibitors plus chemotherapy can benefit patients with metastatic, non-squamous EGFR-sensitive mutation NSCLC (17). Given that duvalizumab is often used as a third-line or later treatment for advanced NSCLC, and the TPS of this patient’s PD-L1 was high (TPS =90%), the patient was switched to a PD-L1 inhibitor (duvalizumab) after experiencing disease progression with osimertinib. However, the efficacy of duvalizumab in this patient was below par. The patient derived minimal benefit from PD-L1 inhibitors, consistent with reports that patients with EGFR driver mutation-positive lung adenocarcinoma have a negative response to immune checkpoint therapy. Yu et al. (18) reported that EGFR exon 19 deletion induces anergic dendritic cells to repress antitumor immunity through exosomes. There is a rich literature available substantiating that patients with EGFR driver mutation have a low expression of PD-L1 (19,20). Interestingly, in this case, the expression of PD-L1 was more than 90%, which may be attributed to individual heterogeneity.
Importantly, this report provides hitherto undocumented evidence of the role of BTN2A1-BRAF in fusion inducing AR to osimertinib. Indeed, the emergence of BRAF fusion may impact the efficacy of osimertinib. Besides, immune checkpoint inhibitors are not recommended for patients with EGFR driver mutations. Our case emphasizes the need for clinicians to conduct a more comprehensive analysis before treatment selection. BRAF inhibitors and/or MEK inhibitors or triple combinations with ERK inhibitors should be considered to overcome resistance to BTN2A1-BRAF.
There were some limitations in this case report. For instance, we did not carry out preclinical experiments, and more radiological evaluations should have been performed after each different treatment. Overall, monotherapy with PD-L1 inhibitors is not indicated according to the TPS of PD-L1 expression, regardless of the mutation of EGFR and other genes.
Conclusions
BTN2A1-BRAF fusion may be a mechanism of resistance to osimertinib, and patients with EGFR driver mutation derive minimal benefit from PD-L1 inhibitors in NSCLC.
Acknowledgments
We thank Jiawen Zheng (Department of Molecular Pathology, The Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital, Zhengzhou, China) for his selfless help. We also thank Home for Researchers (www.home-for-researchers.com) for the linguistic assistance during the preparation of this manuscript.
Funding: This work was supported by the funding from
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2060/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2060/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2060/coif). YJG reports that this study was partly supported by Major Public Welfare Projects in Henan Province [grant number: 201300310400]. CS reports that this study was partly supported by Science and Technology Department of Henan Province [grant number: 212102310128] and Medical science and Technology project of Henan Province [grant number: SBGJ202103033]. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the relatives of the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Ross JS, Wang K, Chmielecki J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881-90. [Crossref] [PubMed]
- Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195-203. [Crossref] [PubMed]
- Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res 2020;26:2654-63. [Crossref] [PubMed]
- Botton T, Talevich E, Mishra VK, et al. Genetic Heterogeneity of BRAF Fusion Kinases in Melanoma Affects Drug Responses. Cell Rep 2019;29:573-88.e7. [Crossref] [PubMed]
- Eisenhardt AE, Sprenger A, Röring M, et al. Phospho-proteomic analyses of B-Raf protein complexes reveal new regulatory principles. Oncotarget 2016;7:26628-52. [Crossref] [PubMed]
- Lu H, Villafane N, Dogruluk T, et al. Engineering and Functional Characterization of Fusion Genes Identifies Novel Oncogenic Drivers of Cancer. Cancer Res 2017;77:3502-12. [Crossref] [PubMed]
- Jang JS, Lee A, Li J, et al. Common Oncogene Mutations and Novel SND1-BRAF Transcript Fusion in Lung Adenocarcinoma from Never Smokers. Sci Rep 2015;5:9755. [Crossref] [PubMed]
- Kim HS, Jung M, Kang HN, et al. Oncogenic BRAF fusions in mucosal melanomas activate the MAPK pathway and are sensitive to MEK/PI3K inhibition or MEK/CDK4/6 inhibition. Oncogene 2017;36:3334-45. [Crossref] [PubMed]
- Vojnic M, Kubota D, Kurzatkowski C, et al. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR therapy in EGFR-Mutated Lung Cancers. J Thorac Oncol 2019;14:802-15. [Crossref] [PubMed]
- Wang CY, Hsia JY, Li CH, et al. Lung Adenocarcinoma With Primary LIMD1-BRAF Fusion Treated With MEK Inhibitor: A Case Report. Clin Lung Cancer 2021;22:e878-80. [Crossref] [PubMed]
- Peng P, Lv G, Hu J, et al. Co-mutations of epidermal growth factor receptor and BRAF in Chinese non-small cell lung cancer patients. Ann Transl Med 2021;9:1321. [Crossref] [PubMed]
- Schrock AB, Zhu VW, Hsieh WS, et al. Receptor Tyrosine Kinase Fusions and BRAF Kinase Fusions are Rare but Actionable Resistance Mechanisms to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol 2018;13:1312-23. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med 2018;378:2288-301. [Crossref] [PubMed]
- Yu S, Sha H, Qin X, et al. EGFR E746-A750 deletion in lung cancer represses antitumor immunity through the exosome-mediated inhibition of dendritic cells. Oncogene 2020;39:2643-57. [Crossref] [PubMed]
- Lu Y, Wu F, Cao Q, et al. B7-H4 is increased in lung adenocarcinoma harboring EGFR-activating mutations and contributes to immunosuppression. Oncogene 2022;41:704-17. [Crossref] [PubMed]
- Negrao MV, Skoulidis F, Montesion M, et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J Immunother Cancer 2021;9:e002891. [Crossref] [PubMed]


