
Expression and clinical significance of undifferentiated embryonic cell transcription factor 1 in breast cancer
Highlight box
Key findings
• UTF1 is closely related to the malignancy and prognosis of breast cancer.
What is known and what is new?
• The common tumor markers still have some limitations in clinical application
• UTF1 has the potential to be a new tumor marker for breast cancer
What is the implication, and what should change now?
• It means that the possibility of targeting UFT1 in the treatment of breast cancer, and it is necessary to find and explore the drugs and mechanisms of targeting UTF1 in the treatment of breast cancer.
Introduction
Breast cancer is the most common cancer in women worldwide, accounting for 30% of all cancer cases. Although diagnostic and treatment strategies for breast cancer have improved significantly over the past few decades, breast cancer remains responsible for 15.5% of cancer-related deaths in women (1). At present, biomarkers such as estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER-2) are widely used to guide clinical treatment and prognostic assessment based on their good predictive effects on treatment sensitivity (2,3). However, due to the heterogeneity of breast cancer, these markers have certain limitations for clinical prognostic evaluation. Therefore, there is an urgent need for new breast cancer biomarkers to guide clinical work (4).
Undifferentiated embryonic cell transcription factor 1 (UTF1), a transcription-related factor, has previously been reported to play a role in embryonic development as well as promoting the reprogramming of human somatic cells into induced pluripotent stem cells (5,6). However, recent studies have found that UTF1 is closely related to the occurrence and development of tumors. Expression of UTF1 in different cancers reveals its function as an oncogene or tumor suppressor, indicating the tissue-specific role of this protein. For example, UTF1 expression is increased in endometrial and prostate cancers and decreased in colon and kidney cancers (7). Moreover, UTF1 expression has potential implications in brain cancer. It can distinguish between grade I–III and IV neuroblastoma and can be used as a good prognostic marker (8).
Functionally, the depletion of UTF1 reduced the tumorigenicity of teratoma, whereas its overexpression resulted in large tumor formation in nude mice, suggesting that UTF1 may have an oncogenic role. This role may explain its ability to promote teratoma formation and its expression in various cancer cells (9,10). In contrast, UTF1 inhibits cell proliferation by directly binding to the promoter of the cell cycle regulator p27 Kip1 to induce cell cycle arrest in cervical cancer, suggesting that UTF1 may also have tumor suppressor activity (11).In this study, we found that UTF1 is closely related to the proliferation, invasion, and metastasis of breast cancer, and has important value in evaluating clinical prognosis, which indicates that UTF1 may be a potential biomarker for breast cancer. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2890/rc).
Methods
Patient
A total of 221 consecutive breast cancer patients who underwent surgical treatment at Shaoxing People’s Hospital from January 2009 to December 2011 were included in this study. The inclusion criteria were as follows: (I) breast cancer confirmed by histopathology; (II) no distant metastasis; (III) newly diagnosed patients who have not received other treatment; (IV) no previous breast surgery history; (V) no malignant lesions; (VI) complete clinical information. The pathological features of breast cancer specimens were classified according to the seventh edition of the Handbook of the International Union against Cancer and the American Joint Committee on Cancer Staging.
Ethical approval
This study was approved by the Ethics Committee of Shaoxing People’s Hospital (No. 2019-K-Y-256-01). All participants or their guardians provided written informed consent or oral informed consent by telephone (recording). The two types of forms of consent were approved by the Ethics Committee of Shaoxing People’s Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Immunohistochemical analysis
Formalin-fixed, paraffin-embedded tissue specimens were used for immunohistochemical (IHC) staining. A PV 2-step IHC kit combined with 3,3'-diaminobenzidine (DAB) kit was used to visualize the reaction products. The primary antibodies used were as follows: anti-UTF1 (#48148, SAB Biotherapeutics, Sioux Falls, SD, USA), Anti-ER (ab32063, Abcam, Cambridge, UK), Anti-PR (ab32085, Abcam, UK), and Anti-Ki67 (ab16667, Abcam, UK).
The IHC evaluation of UTF1, PR, and ER was performed using a semi-quantitative scoring system based on the combination of the proportions of positively stained cells (PS) and the intensity of the staining (IS). The IS was as follows: 0, no staining; 1, weak staining (light yellow); 2, moderate staining (yellow brown); and 3, strong staining (brown). PS ranged from 1% to 100%, given as follows: 0, 0%; 1, 0–25%; 2, 25–50%; 3, 50–75%; 4, 75–100%. Calculation of the 2 scores gave a final score ranging from 0 to 12. Positive expression of UTF1 was defined as the total score ≥1, and positive expression of ER and PR was defined as the total score ≥2.
Evaluation criteria for Ki67 expression were based on the percentage of positive tumor cells. It was as follows: 0, 0%; 1, 0–30%; 2, >30%. A score greater than 1 was defined as Ki67 positive expression.
Follow-up
Telephone and outpatient visits were the main methods of follow-up. Follow up was performed every 3 months within 2 years after operation, once every half a year within 3–5 years after operation, and once a year over 5 years after operation. All patients were followed up until death or the last follow-up date on 30 December 2020. A total of 47 patients (21.2%) died during the follow-up, and the median follow-up interval was 120 months (20–125 months).
Cell culture
Bcap37 was selected for experiments due to its high overexpression and knockdown efficiency. Human breast cancer cell line Bcap37 were purchased from FuHeng Cell Center (FH0216; Shanghai, China). Bcap37 was subcultured in 1640 medium supplemented with 10% fetal bovine serum (FBS) in an incubator at 37 ℃ and 5% CO2 saturation humidity.
Cell transfection
UTF1 overexpressed plasmid (pEGFP-N1-UTF1) and its empty plasmid were purchased from Shanghai Nuoyue Biotechnology Co., Ltd. (Shanghai, China). The pEGFP-N1-UTF1 was transfected into human breast cancer cell line Bcap37 using Lipofectamine 2000 (#11668019, Invitrogen, Carlsbad, CA, USA), and screened by G418 [HY-17561, MedChemExpress (MCE), Monmouth Junction, NJ, USA]. Finally, the cell line BCAP37-UTF1 with stable overexpression of UTF1 was obtained. UTF1 small interfering RNA (siRNA; F: 5-GCUACAGUUCCUUAAAGATT-3; R: 5-UCUUAAGGAACUUGUAGGTT-3), and their matching transfection reagents were purchased from Guangzhou Ruibo Biological Co., Ltd. (Guangzhou, China). According to the manufacturer’s instructions, siRNA was transferred into BCAP37-UTF1 stable cell lines with transfection reagents to down-regulate UTF1 expression.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from logarithmic growth phase cells. RNA was reverse transcribed into complementary DNA (cDNA) by using PrimeScript RT reagent Kit (RR036A, Takara, Shiga, Japan). The qRT-PCR reactions were performed with SYBR® Premix Ex Taq™ Kit (RR820A, Takara, Japan) in LightCycler480 PCR instrument (Roche, Indianapolis, IN, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control. The relative expression of UTF1 was calculated using 2−ΔΔCt method. The primers used for RT-qPCR were as follows: UTF1, forward: 5'-CGCCGCTACAAGTTCCTTAAA-3', reverse: 5'-GGATCTGCTCGTCGAAGG-3'; GAPDH, primer: 5'-CCCAGCAAGAGCACAAGG-3', reverse: 5'-GGTCTACATGGCAACTGTGAGGA-3'.
Western blot
Cells were lysed with a mixture of cell lysis buffer and protease inhibitor. Subsequently, cells were centrifuged at 12,000 r/min at 4 ℃ for 10 minutes. Protein concentration of the supernatant was determined by bicinchoninic acid (BCA) method. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane and sealed with 5% defatted milk powder. Further, the membranes were incubated with UTF1 (#48148, SAB, USA) and β-actin (ab115777, Abcam, UK) primary antibody at 4 ℃ overnight. Membranes were then incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 2 hours. The gel imaging system was used to obtain images after addition of enhanced chemiluminescence (ECL) reagent to the membranes (Biosharp, Hefei, China).
Cell proliferation and colony formation assay
Logarithmic growth phase cells were collected and inoculated into 96-well plates at a density of 5×103 cells/well, each group had 6 parallel holes. Cell viability was tested with the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) by measuring optical density (OD)570 nm every 24 hours for 3 days. The experiment was conducted in 3 independent replicates and all assays were performed in triplicate. In the colony formation assay, the transfected cells were cultured in 6-well plates at a density of 200 cells/well. The culture was terminated when the cell clone was visible to the naked eyes, then fixed cells with 4% paraformaldehyde for 15 minutes followed by 0.1% crystal violet staining for 30 minutes.
Wound healing assay
Bcap37, Bcap37-UTF1, and Si-UTF1 were seeded in 6-well dishes at a density of 1×105 cells per well and grown to 90% confluence. The cell monolayers were scraped with a sterile 10 µL micropipette tip to create a denuded area of constant width. Exfoliated cells were removed by phosphate-buffered saline (PBS) and cultured in the incubator after adding culture medium. The wound closure was monitored and photographed at 24 and 48 hours after wounding.
Migration assay
Migrations assays were performed in 24-well Transwell chamber (Corning, New York, NY, USA). Cells were inoculated at a density of 5×104 cells per well in the upper chamber, and 500 µL medium containing 10% FBS was added in the lower chamber as a chemotactic inducer. After incubation for 48 hours, cells that had migrated to the lower surface of the filter were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet solution.
mRNA sequencing
Total RNA was extracted from Bcap37-UTF1 cells in the logarithmic growth phase and their empty control cells, using TRizol reagent (#15596026, Invitrogen, USA). The RNA samples were sent to Shanghai Xinchao Biotechnology Co., Ltd. (Shanghai, China) for the preparation of the whole transcriptome library and deep sequencing.
Statistical analyses
The statistical software SPSS 26.0 (IBM Corp., Armonk, NY, USA) was used for statistical analyses. Clinical measurement data were expressed as mean ± standard deviation (mean ± SD). T-test and χ2-test were used to evaluate any potential association between UTF1 expression and clinicopathological parameters. Kaplan-Meier method was used to evaluate the survival rate, and the statistical difference between survival curves was determined with the log-rank test. Cox proportional hazard model was used for multivariate prognosis analysis. Variables included in the multivariate analysis were those that were statistically significant in the univariate analysis. All basic experiments were repeated at least three times, and data are presented as the (means ± SD). T-test was used to determine statistical significance between two groups. Statistical significance was considered when P<0.05.
Results
Expression of UTF1 protein in breast cancer
The expression profile of UTF1 in 221 breast cancer tissues was explored by IHC. The results showed that UTF1 was mainly localized in the cytoplasm, and UTF1 was positive in 70 of 221 patients (31.7%) (Figure 1).
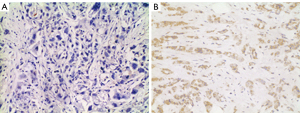
Correlation between UTF1 expression and other clinicopathological parameters in breast cancer
The correlation between UTF1 expression level and clinicopathological parameters of breast cancer (such as patient age, tumor histological stage, tumor size, lymph node metastasis, and expression levels of ER, PR, HER-2, and Ki67), was evaluated (Table 1). The results showed that UTF1 was not correlated with patient age, tumor histological stage, lymph node metastasis, and expression levels of ER, PR, HER-2, and Ki67. However, a significant correlation was observed between tumor size and UTF1 expression level (P=0.004).
Table 1
| Variables | Total patients (n=221) | UTF1 expression | P value | |
|---|---|---|---|---|
| Positive (n=70) | Negative (n=151) | |||
| Age (years) | 221 (100.0%) | 52.8±112.1 | 53.7±10.0 | 0.577 |
| Tumor grade | 0.083 | |||
| I/II | 108 (48.9%) | 28 | 80 | |
| III | 113 (51.1%) | 42 | 71 | |
| Tumor size (cm) | 0.004 | |||
| ≤2 | 117 (52.9%) | 27 | 90 | |
| >2 | 104 (47.1%) | 43 | 61 | |
| Lymph node metastasis | 0.242 | |||
| Negative | 130 (58.8%) | 37 | 93 | |
| Positive | 91 (41.2%) | 33 | 58 | |
| ER | 0.442 | |||
| Negative | 73 (33.0%) | 26 | 47 | |
| Positive | 148 (67.0%) | 44 | 104 | |
| PR | 0.238 | |||
| Negative | 85 (38.5%) | 31 | 54 | |
| Positive | 136 (61.5%) | 39 | 97 | |
| HER-2 | 0.740 | |||
| Negative | 165 (74.7%) | 51 | 114 | |
| Positive | 56 (25.3%) | 19 | 37 | |
| Ki67 | 0.660 | |||
| <30 | 92 (41.6%) | 31 | 61 | |
| ≥30 | 129 (58.4%) | 39 | 90 | |
Data are presented as mean ± SD or n (%). ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor-2.
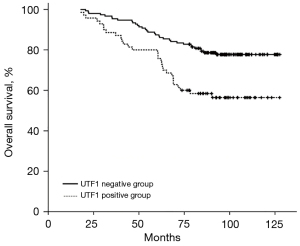
Expression of UTF1 and survival prognosis in breast cancer
Univariate analysis showed that tumor size, ER, PR, lymph node metastasis, and UTF1 expression level were associated with the prognosis of breast cancer (Table 2). Kaplan-Meier analysis was used to obtain the overall survival (OS) time of breast cancer patients from 10-year follow-up and to explore the role of UTF1 in prognosis of breast cancer patients. The results showed that the survival time of cases with positive expression of UTF1 was shorter relative to that of cases with negative expression (Figure 2). Moreover, multivariate Cox risk regression analysis was conducted to evaluate the effect of UTF1 and other clinicopathological parameters on prognosis of breast cancer patients. The findings showed that UTF1 was an independent prognostic factor for predicting the 10-year OS rate of breast cancer patients, with a hazard ratio (HR) value of 1.820 [95% confidence interval (CI): 1.098 to 3.019, P=0.020; Table 2].
Table 2
| Variables | Univariate analysis P value | Multivariate analysis* | |
|---|---|---|---|
| HR (95% CI) | P value | ||
| Lymph node metastasis (positive vs. negative) | <0.001 | 2.530 (1.484–4.311) | 0.001 |
| Tumor size (>2 vs. ≤2) | <0.001 | 2.128 (1.208–3.748) | 0.009 |
| ER (positive vs. negative) | <0.001 | 0.415 (0.252–0.686) | 0.001 |
| UTF1 (positive vs. negative) | 0.001 | 1.820 (1.098–3.019) | 0.020 |
| PR (positive vs. negative) | 0.434 | – | – |
| HER-2 (positive vs. negative) | 0.830 | – | – |
| Ki67 (>30 vs. ≤30) | 0.898 | – | – |
| Age (>40 vs. ≤40) | 0.333 | – | – |
| Tumor grade (III vs. I/II) | 0.548 | – | – |
*, Cox proportional hazards model. ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor-2; HR, hazard ratio; CI, confidence interval.
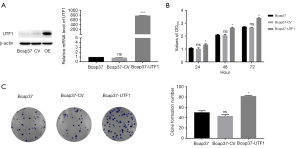
UTF1 promotes growth and proliferation of breast cancer cells
The Bcap37 cell line stably overexpressing UTF1 was constructed in the present study to explore the biological role of UTF1 in breast cancer. Western blot (WB) and RT-qPCR analyses showed that UTF1 expression at the protein and messenger RNA (mRNA) levels were significantly higher in Bcap37 cells transfected with UTF1 compared with the expression level in the control group (Figure 3A). The experimental results indicated that the plasmid transfection efficiency was good, therefore, it was used for subsequent functional experiments. The effect of overexpression of UTF1 on cell proliferation was determined by MTT, and the absorbance was measured at 24, 48, and 72 hours. The results showed that overexpression of UTF1 significantly promoted proliferation rate of Bcap37 cells compared with the control group (Figure 3B). Subsequently, cells from the experimental group and control group were plated on 6-well plate at the same density and at the same time. Cells were stained with crystal violet after observation of visible cell colonies. The staining results showed that the average colony number of Bcap37 cells transfected with UTF1 was significantly higher relative to the colony number of control cells (Figure 3C). The significant difference in colony number between the two groups indicated that UTF1 overexpression promoted cell colony formation and proliferation.
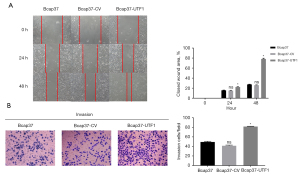
UTF1 promotes migration and invasion of breast cancer cells in vitro
The results reported in the previous sections indicate that UTF1 promotes the growth of breast cancer cells. Therefore, further analyses were performed to explore other biological role of UTF1 in breast cancer cells. A scratch test was conducted to evaluate the effect of UTF1 on Bcap37 cell migration. The percentage of wound healing in the Bcap37-UTF1 group was significantly higher compared with that of the control group (Figure 4A). Subsequently, the role of UTF1 in breast cancer invasion was explored through transwell migration assay. The number of cells that migrated to the matrix glue in the Bcap37-UTF1 group was significantly higher compared with that in the control group (P<0.01; Figure 4B). These findings indicate that UTF1 promotes migration and invasion of breast cancer cells in vitro.
Downregulation of UTF1 inhibits growth and invasion of breast cancer cells
Considering the low expression of UTF1 in BCAP-37 cell line, we envisioned to construct a UTF1 knockdown model based on BCAP37-UTF1 cell line to further verify the role of UTF1 in breast cancer. Firstly, we verified the feasibility of knocking down UTF1 in BCAP37-UTF1 cell line. WB and RT-qPCR results showed that UTF1 protein and mRNA levels were significantly decreased in Si-UTF1 (BCAP37-UTF1 cell line transfected with UTF1 siRNA) compared with BCAP37-UTF1, indicating that UTF1 siRNA can effectively silence the UTF1 gene in the BCAP37-UTF1 stable transgenic cell line (Figure 5A,5B). Therefore, Si-UTF1 was retained for subsequent trials.
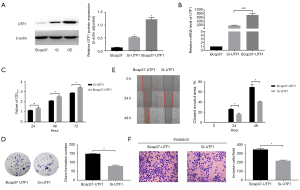
The MTT assay showed that the Si-UTF1 group had a relatively lower rate of proliferation compared with the Bcap37-UTF1 group (Figure 5C). In addition, the colony formation assay showed that the number of colonies formed in the Si-UTF1 group was significantly lower compared with that in the Bcap37-UTF1 group (Figure 5D). This indicates that downregulation of UTF1 inhibited the proliferation ability of breast cancer cells. A wound healing assay and migration assay were performed to further explore the effect of downregulation of the UTF1 gene on migration and invasion of breast cancer cells. The percentage of wound healing in the Si-UTF1 group was significantly lower relative to that in the Bcap37-UTF1 group (Figure 5E). The number of cells that migrated to the matrix in the Si-UTF1 group was significantly lower compared with the number of migrated cells in the Bcap37-UTF1 group (Figure 5F). These results indicate that downregulation of UTF1 inhibits invasion and migration of breast cancer cells in vitro.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differential genes
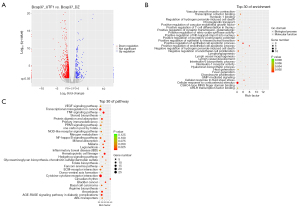
The Bcap37-UTF1 sequence obtained through sequencing experiments was compared with the reference genome. Differences between the two groups were explored using the edgeR software package. The absolute value of fold change (FC) ≥2 and P<0.05 were used as the threshold for the differential genes between the two groups. The top 10 genes with the most significant differences were identified (Table 3). The profile of differentially expressed genes (DEGs) is shown in Figure 6A. The biological roles of DEGs were explored using the GO database (http://www.geneontology.org/) to identify enriched GO terms. GO function analyses showed that the DEGs were implicated in cell apoptotic and transcription cofactor binding (Figure 6B). KEGG is a database for analysis of signaling pathways related to different genes (https://www.kegg.jp/). KEGG signaling pathway analysis showed that the DEGs were associated with cytokine-cytokine receptor interaction and nuclear factor (NF)-kappa B signaling pathway (Figure 6C).
Table 3
| Gene ID | Gene name | Bcap37_UTF1_count | Bcap37_CV_count | log2FC | P value | Up/down |
|---|---|---|---|---|---|---|
| ENSG00000198888 | MT-ND1 | 1 | 75950 | −16.14 | 0.0000 | Down |
| ENSG00000198763 | MT-ND2 | 54079 | 153 | 8.54 | 0.0000 | UP |
| ENSG00000211459 | MT-RNR1 | 148 | 11492 | −6.21 | 0.0000 | Down |
| ENSG00000255508 | RP11-864I4.1 | 3169 | 133 | 4.65 | 0.0000 | UP |
| ENSG00000111432 | FZD10 | 1884 | 81 | 4.61 | 0.0000 | UP |
| ENSG00000100092 | SH3BP1 | 41 | 1041 | −4.59 | 0.0000 | Down |
| ENSG00000213443 | RP11-75L1.2 | 555 | 18 | 5.02 | 0.0000 | UP |
| ENSG00000241360 | PDXP | 847 | 69 | 3.69 | 0.0000 | UP |
| ENSG00000263020 | XXbac-BPG32J3.22 | 1067 | 117 | 3.26 | 0.0000 | UP |
| ENSG00000213240 | RP11-458D21.5 | 187 | 1 | 7.62 | 0.0000 | UP |
Discussion
At present, there are still certain limitations for traditional breast cancer biomarkers. One such an example is Ki-67, although it has shown certain clinical prognostic value, its poor inter-laboratory reproducibility and the difference in threshold between low proliferation index and high proliferation index make its clinical application still controversial (12). For ER, the unavoidable problem is the close crosstalk between ER and HER family signaling pathways, which is not only the basic factor of resistance to endocrine therapy targeting the ER pathway, but also one of the reasons for the bias of ER in clinical prognosis evaluation (13). In addition, a recent study reported that the prognostic value can be improved by combining ER, PR, Ki-67, and HER2 (14), but a subsequent study found that this prognostic evaluation tool still cannot cover various subtypes well (15). Therefore, new proteomic markers are urgently needed to further improve this dilemma.
In this study, we found that the poor prognosis of breast cancer and the malignant biological characteristics of breast cancer cells were correlated with the high expression of UTF1, which plays a role as an oncogenic factor in breast cancer. However, this is in contrast to the results reported previously, which suggested that UTF1 expression is down-regulated in breast cancer and that UTF1 functions as a protective factor in breast tissue (16). It is worth noting that this paradox is also present in cervical cancer (11,17). As for this phenomenon, as with most transcription factors, it may be related to the dual role of UTF1 in carcinogenesis. We believe that the possible explanation is that in addition to the extensive tissue specificity, UTF1 has certain subtype specificity in both breast and cervical cancer. Further subtype analysis is needed to confirm this. In addition, there are several other viewpoints, as follows. Ethnic variation is at the forefront of the problem, as different studies enrolled patients of different ethnic groups, which may affect the results. Second, different/unclear cut-off values have been used in UTF1 expression analyses. For example, in one study, the median was used as the basis for determining the cut-off point (18), whereas in other similar studies, the mean value or even positive or negative expression of genes was used for this purpose. Third, sample size is a key factor affecting the association between UTF1 expression and the pathological features and prognosis of BC.
A previous study showed that sample size affects the results of microarray analysis, and small sample size can lead to unstable gene lists and poor prediction accuracy (19). Future studies using larger cohorts of patients are needed to validate our findings.
In addition, by sequencing analysis, we also found that UTF1 was not only closely related to apoptosis genes, but also closely related to the NF-kappa B pathway. The close link between the NF-kappa B pathway and apoptosis has long been demonstrated in multiple experiments (20-22). Uncontrolled regulation of the NF-kappa B pathway can lead to uncontrolled cell growth. Therefore, we suggest that UTF1 may promote breast cancer progression by regulating NF-kappa B signaling to affect cell apoptosis. However, further experiments are needed to verify this.
In summary, this study systematically verified the close relationship between UTF1 and breast malignant behavior from multiple perspectives, and verified the association between UTF1 and clinical prognosis. It is a potential biomarker of breast cancer, which not only enriches the subtype grouping of breast cancer, but also improves the prognostic algorithm, so as to assist clinical treatment more effectively. However, this study also had some limitations. First, as a retrospective study, this study was susceptible to selection bias. Secondly, the small sample size of this study and the small number of terminal events in the survival analysis limit the reliability of the results. Third, there is heterogeneity in adjuvant chemotherapy regimens, which affects the study results. Finally, due to the low expression level of UTF1 and the lack of standard staining protocols and analytical methods, the reliability and accuracy of its IHC staining need to be verified. Therefore, large samples, standardized clinical data, and more in-depth mechanism exploration will be the directions of our next efforts.
Conclusions
Our study shows that UTF1 is closely related to the malignancy and prognosis of breast cancer, suggesting the potential of UTF1 as a breast cancer biomarker, and that the NF-kappa B pathway as a potential UTF1 signaling pathway, will be the focus of future research.
Acknowledgments
Funding: This study was supported by the Jinhua Science and Technology Research Program (No. 2021-3-084), the Jinhua central hospital non-profit technology applied research of Zhejiang China (No. JY2020-5-04), and the Public Welfare Projects of Jinhua Science and Technology Bureau, Zhejiang Province (No. 2018-4-016).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2890/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2890/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2890/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shaoxing People’s Hospital (No. 2019-K-Y-256-01). All participants or their guardians provided informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Du L, Yau C, Brown-Swigart L, et al. Predicted sensitivity to endocrine therapy for stage II-III hormone receptor-positive and HER2-negative (HR+/HER2) breast cancer before chemo-endocrine therapy. Ann Oncol 2021;32:642-51. [Crossref] [PubMed]
- Sueta A, Yamamoto Y, Hayashi M, et al. Clinical significance of pretherapeutic Ki67 as a predictive parameter for response to neoadjuvant chemotherapy in breast cancer: is it equally useful across tumor subtypes? Surgery 2014;155:927-35. [Crossref] [PubMed]
- Paizula X, Mutailipu D, Xu W, et al. Identification of biomarkers related to tumorigenesis and prognosis in breast cancer. Gland Surg 2022;11:1472-88. [Crossref] [PubMed]
- Okuda A, Fukushima A, Nishimoto M, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J 1998;17:2019-32. [Crossref] [PubMed]
- Schwarz BA, Cetinbas M, Clement K, et al. Prospective Isolation of Poised iPSC Intermediates Reveals Principles of Cellular Reprogramming. Cell Stem Cell 2018;23:289-305.e5. [Crossref] [PubMed]
- Mouallif M, Albert A, Zeddou M, et al. Expression profile of undifferentiated cell transcription factor 1 in normal and cancerous human epithelia. Int J Exp Pathol 2014;95:251-9. [Crossref] [PubMed]
- Melone MA, Giuliano M, Squillaro T, et al. Genes involved in regulation of stem cell properties: studies on their expression in a small cohort of neuroblastoma patients. Cancer Biol Ther 2009;8:1300-6. [Crossref] [PubMed]
- Nishimoto M, Miyagi S, Yamagishi T, et al. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol Cell Biol 2005;25:5084-94. [Crossref] [PubMed]
- Jia J, Zheng X, Hu G, et al. Regulation of pluripotency and self- renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell 2012;151:576-89. [Crossref] [PubMed]
- Wu XL, Zheng PS. Undifferentiated embryonic cell transcription factor-1 (UTF1) inhibits the growth of cervical cancer cells by transactivating p27Kip1. Carcinogenesis 2013;34:1660-8. [Crossref] [PubMed]
- Duffy MJ, Harbeck N, Nap M, et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer 2017;75:284-98. [Crossref] [PubMed]
- Arpino G, Wiechmann L, Osborne CK, et al. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev 2008;29:217-33. [Crossref] [PubMed]
- Cuzick J, Dowsett M, Pineda S, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol 2011;29:4273-8. [Crossref] [PubMed]
- Harris LN, Ismaila N, McShane LM, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 2016;34:1134-50. [Crossref] [PubMed]
- Xu C, Zhou Y, Chen W. Expression of undifferentiated embryonic cell transcription factor-1 (UTF1) in breast cancers and their matched normal tissues. Cancer Cell Int 2014;14:116. [Crossref] [PubMed]
- Guenin S, Mouallif M, Deplus R, et al. Aberrant promoter methylation and expression of UTF1 during cervical carcinogenesis. PLoS One 2012;7:e42704. [Crossref] [PubMed]
- Barros-Oliveira MDC, Costa-Silva DR, Campos-Verdes LC, et al. CYP19A1 gene expression in the peripheral blood of Brazilian women with breast cancer relapse. BMC Cancer 2020;20:480. [Crossref] [PubMed]
- Stretch C, Khan S, Asgarian N, et al. Effects of sample size on differential gene expression, rank order and prediction accuracy of a gene signature. PLoS One 2013;8:e65380. [Crossref] [PubMed]
- Qiu J, Zhang T, Zhu X, et al. Hyperoside Induces Breast Cancer Cells Apoptosis via ROS-Mediated NF-κB Signaling Pathway. Int J Mol Sci 2019;21:131. [Crossref] [PubMed]
- Chen M, Xiao C, Jiang W, et al. Capsaicin Inhibits Proliferation and Induces Apoptosis in Breast Cancer by Down-Regulating FBI-1-Mediated NF-κB Pathway. Drug Des Devel Ther 2021;15:125-40. [Crossref] [PubMed]
- Zeng A, Liang X, Zhu S, et al. Chlorogenic acid induces apoptosis, inhibits metastasis and improves antitumor immunity in breast cancer via the NF-κB signaling pathway. Oncol Rep 2021;45:717-27. [Crossref] [PubMed]
(English Language Editor: J. Jones)


