
Acquired BRAF gene fusions in Osimertinib resistant EGFR-mutant non-small cell lung cancer
BRAF kinase somatic missense mutations, identified in up to 66% of malignant melanomas (1) and in 6–8% of non-small cell lung cancers (NSCLCs) (2), cause hyperactivation of the mitogen-activated protein kinase (MAPK) pathway. In a similar way, BRAF oncogenic fusion proteins also activate MAPK signaling independently of RAS activation (Figure 1). Moreover, the differential subcellular localization of BRAF protein is controlled by the N-terminal fusion partner and the physical localization into cell can regulate the specific oncogenic mechanisms, similarly to the effect observed in other fusion oncoproteins (3). Chimeric oncoproteins often contain the C-terminal kinase domain of the RTK joined in cis to various N-terminal non-kinase fusion partners. In ROS1 kinase fusion oncoproteins, it was previously shown that distinct N-terminal fusion partners drive differential subcellular localization with specific signaling effects: SCD4-ROS1 and SLC34A2-ROS1 fusion oncoproteins lie on endosomes, activating the MAPK pathway, while CD74-ROS1 variants are seen in the endoplasmic reticulum with impaired MAPK activation (3). Interestingly, forced relocalization of CD74-ROS1 from the endoplasmic reticulum to endosomes restored MAPK signaling. Moreover, ROS1 fusion oncoproteins, that better activate MAPK, developed more aggressive tumors. Therefore, importantly, differential subcellular localization that could be controlled by the N-terminal fusion partner regulates the oncogenic mechanisms and output of some fusion oncoproteins (3). BRAF fusion genes are described in several tumor types with a broad variety of 5’ fusion partners (more than 60 different partners reported). In addition, it has been seen that the 5’ fusion partners can redirect the subcellular localization of the BRAF fusion proteins, therefore, also modulating the intensity by which BRAF kinase domain can activate downstream MEK (4). In patient derived colorectal cancer organoids it was noted that the subcellular localization of the BRAF fusion protein, as well as the level of MAPK pathway activation, were regulated by the 5’ partner. The localization of DLG1-BRAF proteins at the plasma membrane could confer enhanced activation of the MAPK pathway compared with AGAP3-BRAF, TRIM24-BRAF, and the canonical BRAFV600E mutation that exhibits a diffused localization pattern throughout the cytoplasm (4). Wild-type BRAF is expressed throughout the cytoplasm and is recruited to the plasma membrane through activated RAS. Redirected subcellular localization by the 5’ fusion partners influence the function of oncogenic BRAF and raise the interest of using 3D tumor organoids in assessing intracellular signaling and sensitivity to targeted therapies of diverse BRAF fusion genes (4). BRAF fusion genes are also implicated in resistance to osimertinib in EGFR-mutant NSCLC (5). In this issue of Translational Cancer Research, Kong and colleagues report the case of a woman with lung adenocarcinoma harboring EGFR exon 19 deletion who received later-line osimertinib and developed a novel butyrophilin subfamily 2 member A1 (BTN2A1)-BRAF fusion that was refractory to duvalizumab (an anti-PD-L1 antibody) (6).
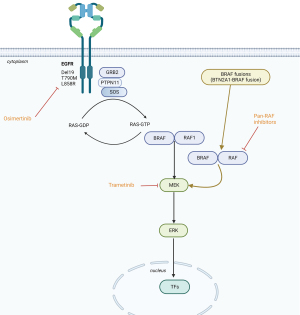
What is known about BRAF fusions in NSCLC? BRAF is modified in 4.5% of patients analyzed [1,048 of 23,3976 (7), in which37% are BRAF V600E, 38% are non-V600E, and 18% are BRAF inactivating, kinase-dead, mutations]. Rearrangements have been seen in 4%, consisting of N-terminal deletion (0.8%), kinase domain duplications (0.8%), and BRAF fusions (3%) (7). The staphylococcal nuclease and tudor domain containing 1 (SND1)-BRAF fusion has been seen three times, while armadillo repeat containing 10 (ARMC10), dedicator of cytokinesis 4 (DOCK4), and tripartite motif containing 24 (TRIM24)-BRAF were each identified twice (7). Seventeen other BRAF fusions harbored separate partners, again combining the kinase domain of BRAF with the N-terminal segment of a distinctive gene product. Henceforth, omission of an N-terminal RAS binding auto-inhibitory domain produced RAS-independent BRAF dimerization (5). Vojnic et al. (8) identified four patients (2.3%) with a BRAF fusion [three with acylglycerol kinase gene (AGK)-BRAF and one with praja ring finger ubiquitin ligase 2 (PJAZ2)-BRAF] in re-biopsies after acquired resistance to EGFR tyrosine kinase inhibitors. Noteworthy was the fact that induction of AGK-BRAF fusion in H1975 (L858R+ T790M), PC9 (ex0n 19 deletion) and HCC827 (exon 19 deletion) cells augmented the phosphorylation of BRAF, MEK1/2, ERK1/2, and signal transducer and activator of transcription 3 (STAT3), causing resistance to growth inhibition by osimertinib (8). BRAF rearrangements are also insensitive to first generation BRAF inhibitors (vemurafenib, dabrafenib or encorafenib) owing to homodimerization and heterodimerization with other RAF proteins. Of interest is the fact that co-treatment of osimertinib with trametinib (MEK inhibitor) abolished tumor growth, as well as the pan-RAF inhibitor (LY3009120) as a single agent blocked growth of all cell lines with mutant EGFR and BRAF fusion (8), suggesting therapeutic alternatives for this mechanisms of resistance in EGFR-mutant cancer.
Schrock et al. (9) analyzed a large set of EGFR-mutated NSCLCs and identified 10 tumors with concurrent BRAF fusions: 7 with EGFR L858R, and 3 with EGFR exon 19 deletion. Fusion partner genes of BRAF included TRIM24, Arf-GAP with GTPase domain, ankyrin repeat and PH domain 3 (AGAP3), AGK, ARMC10, spalt like transcription factor 2 (SALL2), DOCK4, epidermal growth factor receptor pathway substrate 15 (EPS15), growth hormone receptor (GHR), and KIAA1549 (9). Mitsudomi poses the question: How do we treat EGFR-mutant NSCLCs with acquired BRAF fusion? (5). As above mentioned, the combination of osimertinib plus trametinib (MEK inhibitor) has demonstrated relevant synergism (8). Vojnick et al. (8) also demonstrated that a single agent pan-RAF inhibitor (LY3009120) blocked cell line growth with mutant-EGFR and BRAF fusions. Therefore, the combinatory therapy approaches in EGFR-mutant NSCLCs with concurrent acquired BRAF fusions warrant considerable preclinical research. More recent studies also detected TRIM24-BRAF and AGK-BRAF fusions identified with first line osimertinib or with later-line osimertinib (10) as is the case reported here with the novel BTN2A1-BRAF fusion (6). Other BRAF fusions have recently been described in EGFR-mutant osimertinib resistant NSCLC and are related to apolipoprotein b mRNA editing enzyme, catalytic polypeptide-like (APOBEC) mutational signature (11).
The evolutionary trajectory of the multi-therapy EGFR-mutant NSCLC patient following initial treatment with gefitinib epitomizes the clinical situation of EGFR-mutant NSCLC and explains the interest in performing sequential molecular assessment due to the paradigm finding that, after a later line of osimertinib, the co-existence of EGFR exon 19 deletion, T790M, and BTN2A1-BRAF fusions, plus other genetic alterations, occur with lack of response to anti PD-1/PD-L1 antibodies despite high PD-L1 expression (6,8), which is probably enhanced by the alteration in the BTN2A1 expression, and is necessary in cancer cells to empower T cell antitumoral responsiveness (12).
Moreover, although BRAF fusions eliminate the first eight exons of BRAF, promoting BRAF dimerization independent of activated RAS (rat sarcoma viral oncogene homologues), their role as mechanisms of resistance to RAS inhibiting drugs has also been described (13). AKAP9-BRAF fusion, as well as EML4-ALK and CDDC6-RET fusions, have been detected as mechanisms of acquired resistance to adagrasib treatment in KRAS G12C mutant cancers (14).
NSCLCs in never and light smokers often harbor fusions in RAS pathway genes. A gene rearrangement has been identified by RNA sequencing that predicted to encode an N-terminal segment of the tight junction transmembrane protein occluding (OCLN) fused in-frame with a C-terminal segment of the guanine exchange factor (GEF) domain of the RAS activator RASGRF1(OCCLN-RASGRF1) (15), implicating that RAF-MEK-ERK inhibition should be taken into account for therapeutic strategies (16). RASGRF1 encodes a GEF which releases GDP from RAS, allowing to bind GTP and activate MAPK signaling. In addition, another RAS-activating TMEM87A-RASGRF1 fusion was identified by bulk RNA-sequencing in a never-smoker patient with a partial response of 33 months to the multi-kinase inhibitor sunitinib (17).
Although the co-occurrence or acquisition of BRAF fusion in EGFR-mutant and KRAS-mutant patients is uncommon, it unveils a new mechanism of resistance. Whether such co-occurring BRAF fusions in NSCLCs are tractable will require further endeavors to target, such as with pan-RAF inhibitor (18) (Figure 1) or other combinatory approaches to circumvent signaling pathways that occur frequently in EGFR-mutant NSCLCs involving MET and FOXM1 (19).
Acknowledgments
Funding: This work was partially supported by
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2888/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [Crossref] [PubMed]
- Lin S, Lin Y, Nery JR, et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 2014;111:17224-9. [Crossref] [PubMed]
- Neel DS, Allegakoen DV, Olivas V, et al. Differential Subcellular Localization Regulates Oncogenic Signaling by ROS1 Kinase Fusion Proteins. Cancer Res 2019;79:546-56. [Crossref] [PubMed]
- Stangl C, Post JB, van Roosmalen MJ, et al. Diverse BRAF Gene Fusions Confer Resistance to EGFR-Targeted Therapy via Differential Modulation of BRAF Activity. Mol Cancer Res 2020;18:537-48. [Crossref] [PubMed]
- Mitsudomi T. BRAF Fusion - Another Mechanism of Acquired Resistance to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol 2019;14:764-5. [Crossref] [PubMed]
- Kong WM, Guo YJ, Ma J, et al. BTN2A1-BRAF fusion may be a novel mechanism of resistance to osimertinib in lung adenocarcinoma: a case report. Transl Cancer Res 2023;12:186-93. [Crossref] [PubMed]
- Sheikine Y, Pavlick D, Klempner SJ, et al. BRAF in Lung Cancers: Analysis of Patient Cases Reveals Recurrent BRAF Mutations, Fusions, Kinase Duplications, and Concurrent Alterations. JCO Precis Oncol 2018;2:PO.17.00172.
- Vojnic M, Kubota D, Kurzatkowski C, et al. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR therapy in EGFR-Mutated Lung Cancers. J Thorac Oncol 2019;14:802-15. [Crossref] [PubMed]
- Schrock AB, Zhu VW, Hsieh WS, et al. Receptor Tyrosine Kinase Fusions and BRAF Kinase Fusions are Rare but Actionable Resistance Mechanisms to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol 2018;13:1312-23. [Crossref] [PubMed]
- Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res 2020;26:2654-63. [Crossref] [PubMed]
- Selenica P, Marra A, Choudhury NJ, et al. APOBEC mutagenesis, kataegis, chromothripsis in EGFR-mutant osimertinib-resistant lung adenocarcinomas. Ann Oncol 2022;33:1284-95. [Crossref] [PubMed]
- Cano CE, Pasero C, De Gassart A, et al. BTN2A1, an immune checkpoint targeting Vγ9Vδ2 T cell cytotoxicity against malignant cells. Cell Rep 2021;36:109359. [Crossref] [PubMed]
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- Awad MM, Liu S, Rybkin II, et al. Acquired Resistance to KRAS(G12C) Inhibition in Cancer. N Engl J Med 2021;384:2382-93. [Crossref] [PubMed]
- Hunihan L, Zhao D, Lazowski H, et al. RASGRF1 Fusions Activate Oncogenic RAS Signaling and Confer Sensitivity to MEK Inhibition. Clin Cancer Res 2022;28:3091-103. [Crossref] [PubMed]
- Zhu LY, Yuan JB, Zhang L, et al. Loss of MLL Induces Epigenetic Dysregulation of Rasgrf1 to Attenuate Kras-Driven Lung Tumorigenesis. Cancer Res 2022;82:4153-63. [Crossref] [PubMed]
- Cooper AJ, Kobayashi Y, Kim D, et al. Identification of a RAS-activating TMEM87A-RASGRF1 Fusion in an Exceptional Responder to Sunitinib with Non-Small Cell Lung Cancer. Clin Cancer Res 2020;26:4072-9. [Crossref] [PubMed]
- Zhang C, Bollag G. Triple Therapy to Outwit the BRAF Oncogene. Cancer Discov 2021;11:1620-2. [Crossref] [PubMed]
- Rosell R, Cardona AF, Arrieta O, et al. Coregulation of pathways in lung cancer patients with EGFR mutation: therapeutic opportunities. Br J Cancer 2021;125:1602-11. [Crossref] [PubMed]


