
Identification and evaluation of a risk model predicting the prognosis of breast cancer based on characteristic signatures
Highlight box
Key findings
• A predictive prognosis model based on characteristic gene sets of breast cancer was established to predict the survival time of breast cancer patients using a cluster of 12 differentially expressed genes.
What is known and what is new?
• Several biomarkers and models have been reported and established for the prognosis of breast cancer, but their efficacy is underestimated.
• A novel prognosis model based on characteristic gene sets of breast cancer has been introduced.
What is the implication, and what should change now?
• This model could be used as a tool for the prognosis of patients with breast cancer, and further extensive functional exploration revealed the model was closely related to tumor immune microenvironment, especially M2 macrophage infiltration.
Introduction
Breast cancer (BC) is one of the most common types of cancers worldwide and one of the leading fatal cancers in females, accounting for approximately 14% of all cancer-related deaths (1). While BC is a rare disease in males with a much lower rate of incidence, the mortality rate of BC male patients is quadruple that of female patients, making it worth attention (2). According to statistics, there were 194,280 new BC cases in the US in 2009 and 40,610 BC-related deaths (3), while a decade later, those numbers increased to 268,670 and 41,400, respectively (2).
Advancement in the diagnosis of BC may contribute to the rising incidence of BC. Self-inspection of the breasts at an early stage is considered to be effective for the discovery and diagnosis of BC and can prevent BC-related deaths, but there are still numerous BC cases that are only diagnosed at advanced stages due to patient (female) negligence of the importance of self-inspection and regular clinical examination of the breasts. Screening examinations are gradually becoming routine for BC diagnosis, including ultrasound breast imaging, single photon emission computerized tomography, and digital mammography. Mammography is considered the gold standard test for early BC detection (3), although there are concerns, such as radical harm caused by frequent screening examinations, false-positive results and overdiagnosis, which lead to a negative impact on patient quality of life (4). Biomarkers of BC are also used in diagnosis during different stages, including immunohistochemical markers [ER, PR, HER2 (ERBB2), and proliferation marker protein Ki-67 (MKI67)], genomic markers (BRCA1, BRCA2, and PIK3CA), and immunomarkers (tumor-infiltrating lymphocytes and PD-L1) (5). While there has been progress in BC treatment, the mortality rate is not declining, indicating that new biomarkers or models for BC survival prediction and prognosis should be established to help provide more efficient and personalized measurements for managing BC. By 2022, the incidence and mortality of BC in the Chinese population has been predicted to be higher than those in developed countries (6). Given the lack of an effective survival prediction model for physicians, novel biomarkers or models for BC survival prediction and prognosis should be established to help provide more efficient and personalized measures for managing BC in the early stage.
In our study, we identified two characteristic gene sets of BC development, the BC unfavorable gene set and the BC favorable gene set. Survival analysis and ROC analysis were used to determine the diagnostic and prognostic capabilities of the two gene set model scores, respectively. Both the unfavorable and favorable BC gene set models presented reliable biomarkers for BC diagnosis and independent biomarkers for prediction and prognosis. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2444/rc).
Methods
Datasets
The gene expression profiles of BC and healthy breast samples were obtained from The Cancer Genome Atlas (TCGA), including 173 stage I BCs, 597 stage II BCs, 240 stage III BCs, 20 stage IV BCs and 113 normal samples. TCGA is an international database that is publicly accessible and freely available for research and contains a large collection of human cancer genome sequencing data. Previous studies using a group of genes with known alterations in BC to validate TCGA data suggested that TCGA had accurately documented the genomic abnormalities of multiple malignancies (7-9). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of differentially expressed genes (DEGs)
Preliminary identification of the characteristic genes promoting or inhibiting the development of BC was performed using differential expression analysis and Short Time-series Expression Miner (STEM) analysis. STEM is a new software package for analyzing short time-series expression data. The software can find statistically significant patterns from short time-series microarray experiments and compare datasets across experiments. STEM presents its analysis of the data in a highly visual and interactive manner (10). STEM’s unique algorithm tends to cluster and compare short time-series gene expression data combined with its visualization capabilities and integration with Gene Ontology, suggesting that STEM is useful in the analysis of data from a significant portion of all microarray studies (10).
The RNA sequencing expression profile of BC was displayed as read counts, which were subsequently normalized by the voom function in the limma package. P<0.01 adjusted by the false discovery rate (FDR) and |log fold change (FC)| >1.5 were set as thresholds. In the development of BC, if a DEG was gradually upregulated (log FC stage I vs. control < log FC stage II vs. control < log FC stage III vs. control < log FC stage IV vs. control) or gradually downregulated (log FC stage I vs. control > log FC stage II vs. control > log FC stage III vs. control > log FC stage IV vs. control), then it was considered to be a characteristic BC development gene.
Limma package
DEGs in the four stages of BC were identified separately using the limma package with RNA sequencing expression profiles of BC in TCGA. The package can now perform both differential expression and differential splicing analyses of RNA sequencing (RNA seq) data. This software provides an integrated data analysis solution, using advanced computational algorithms to deliver reliable performance on large datasets, represent expression data and simplify the user interface (11). Significantly DEGs were selected by using the FDR and log (FC) values. In the development of BC, if the DEGs were gradually upregulated [log (FC) value increased] or gradually downregulated [log (FC) decreased], they were considered to promote or suppress BC development.
Cox regression analysis
Univariate Cox regression analysis was used to identify genes associated with BC prognosis. Multivariate Cox regression analysis was used to test the correlation among our models, current clinicopathological features and BC prognosis.
Least Absolute Shrinkage and Selection Operator (LASSO) regression analysis
We screened the expression levels for significant genes in the favorable and unfavorable groups using univariate Cox regression analysis (P<0.01). Then, these genes were selected for further functional investigation, and a possible risk score was developed using the LASSO Cox regression algorithm. The most refined prognostic prediction models were constructed by minimal compression using LASSO regression analysis based on two characteristic gene sets of BC development. With a constraint condition in the sum of the absolute terms, LASSO could reduce the dimension of the independent variable and produce a better model fit. It was suitable for analyzing the genetic survival data, identifying more meaningful independent variables and determining the model. The fixed coefficient R2 indicates the fraction of the variation that the covariate explains. The larger the percentage is, the better the model fit (12).
Calculation of characteristic BC development Gene Set Variation Analysis (GSVA) Score
Both models based on upregulated and downregulated characteristic gene sets of BC development were scored. GSVA is a popular method of scoring individual samples for molecular characteristics or gene sets. The GSVA package in R was used to calculate the BC-unfavorable GSVA score and BC-favorable GSVA score for individual samples.
We screened the expression levels for genes in the TCGA data group using univariate Cox regression analysis. Thus, we identified genes that were highly related to survival (P<0.01). We chose these genes for further functional investigation and developed a possible risk score using the LASSO Cox regression algorithm. In detail, the risk score of the TCGA training set was calculated with the linear combination of the signature gene expression weighted by their regression coefficients. Risk score = (exprgene1 × coefficientgene1) + (exprgene2 × coefficientgene2) + … + (exprgenen × coefficientgenen). Finally, minimum criteria defined genes and their constants, choosing the perfect penalty parameter λ related to the minimum 10-fold cross-validation in the training set.
Statistical analysis
Receiver operating characteristic (ROC) curve analysis and survival analysis were used to explore the diagnostic and prognostic capabilities of the two scoring systems. In ROC curve analysis, the area under the curve provides an objective parameter of the diagnostic or prognostic accuracy of a test, which is superior to comparing single combinations of sensitivity and specificity values since the influence of the threshold value is eliminated. Because only part of the ROC curve represents clinically relevant combinations of sensitivity and specificity, comparing the ROC curves in the relevant sensitivity or specificity ranges is preferred over comparing the total area under the curve. In addition, ROC analysis can be used to determine the optimal threshold value for tests that generate continuous quantitative data (13). Multivariate Cox regression analysis was used to compare the relative prognostic value of the two scoring systems with that of routine clinicopathological features. Moreover, correlation analysis was used to explore the correlation between these models and the tumor immune microenvironment and drug responses.
Results
Identification of gradually up/downregulated genes
Compared to normal breast tissue samples, a total of 5,770 DEGs were identified in stage I BCs, 6,083 DEGs in stage II BCs, 5,966 DEGs in stage III BCs, and 5,855 DEGs in stage IV BCs, with a total of 4,656 common DEGs among all stages (Figure 1A,1B). We summarized 10 major variation tendencies of gene expression by STEM analysis in BC gene expression profiles in TCGA (Figure S1). Based on these tendencies, DEGs were classified into two groups: one group (BC1 cluster) contained 476 genes that were gradually upregulated with the development of BC from stage I to stage IV (Figure 1C,1D), indicating the progression of BC; this group was named the BC unfavorable gene set. The second group (BC2 cluster) contained 262 genes that were gradually downregulated with the development of BC (Figure 1E,1F), which indicated a good prognosis, named the BC favorable gene set.

To identify BC DEGs associated with prognosis, we first used univariate Cox regression analysis to assess the association between the expression levels of each DEG and found that 26 genes were significantly associated with BC prognosis in the BC unfavorable gene set (Table S1; P<0.01) and 14 genes in the BC favorable gene set (Table S2; P<0.01).
Establishment and evaluation of a prognostic model based on BC DEGs
LASSO regression analysis was performed to establish risk models concerning BC prediction and prognosis based on the two characteristic gene sets of BC development. The results showed that a total of 12 DEGs (Figure 2A-2D) in the BC unfavorable gene set and 7 DEGs (Figure 2E-2H) in the BC favorable gene set were significantly associated with BC prognosis. On this basis, two risk models were obtained, consisting of 12 genes and 7 genes, named the BC unfavorable model (BC1 model) and BC favorable model (BC2 model), respectively.

ROC analysis was performed to evaluate the prognostic efficacy of the two risk models. In the BC1 model, the AUC was 0.80 (Figure 2I), presenting great capability of the model for BC prognosis, while in the BC2 model, the AUC was 0.66 (Figure 2J). Compared with the BC2 model, the BC1 model was more efficient in prognostic capability. ROC analysis of BC patients at 1, 2, and 3 years after diagnosis was also performed, and the AUCs were 0.80, 0.77, and 0.75, respectively, in the BC1 model (Figure 2K) and 0.66, 0.69, and 0.74 in the BC2 model (Figure 2L). The results showed that both the BC1 and BC2 models could accurately predict the survival rate of BC patients, especially the BC1 model.
Additionally, the efficacy of the risk score, age, stage, T stage, M stage and N stage for BC prognosis was compared. In the BC1 model, the AUC of the risk score was 0.80, which was the highest among these clinical characteristics, illustrating that the BC1 model was more efficient than other clinical factors for prognosis (Figure 2M). In the BC2 model, the AUC of the risk score was 0.64, which was lower than that of the clinical characteristics, including 0.71 for age, 0.64 for stage, 0.70 for T stage, 0.55 for M stage and 0.62 for N stage (Figure 2N).
Survival analysis was performed to evaluate the survival possibility of BC patients divided into a high-risk group and a low-risk group based on these two models. In the BC1 model, the results of survival analysis showed that the survival possibility of the high-risk group was lower than that of the low-risk group, and the gap between the two groups became much wider as the survival years increased (P<0.001; Figure 3A). The final survival time of the high-risk group was less than 22 years, which was shorter than that of the low-risk group (23.5 years). In the scatter graphs, we ranked BC patients by the risk scores evaluated with our model from low scores to high scores on the horizontal axis. The vertical axis represented their survival time; green scatter represents living patients, and red represents patients who died. The graphs demonstrate that more patients with high risk scores died, and more patients with low risk scores were living. The survival time of patients with low risk scores was longer than that of patients with high risk scores. As shown in Figure 3B, the survival time in the high-risk group was significantly lower than that in the low-risk group using the BC1 model. In the BC2 model, the results were similar (Figure 3C,3D), indicating that these two models can accurately predict the survival rate of BC patients.

Association of risk score from BC models with clinical characteristics
Multivariate Cox regression analysis and univariate Cox regression analysis were used to identify factors independently associated with BC prognosis. For both the BC1 and BC2 models, the P values for age, N stage and risk score were all less than 0.001, suggesting that age, lymphatic metastasis and the risk score were all independently associated with BC prognosis. These findings demonstrated that the risk score derived from our model was an independent prognostic predictor of BC patients (Figure 3E-3H).
The performance of the risk scores was tested with each clinical characteristic. Significant differences were observed between risk score and age (P<0.0001; Figure 3I) in the BC1 model, while risk score and tumor topography were significantly correlated (P<0.001; Figure 3J) in the BC2 model.
In the heatmap, there were obviously more patients (≥60 years) with high risk scores, yet there was no distinct regularity between risk scores and other features, which was consistent with the results of multivariate Cox regression analysis that age was an independent factor correlated with the BC1 model (Figure 3I). In the BC2 model, a marked difference was observed between tumor topography and the risk score (Figure 3J).
Immune infiltration analysis and drug susceptibility
To further explore the immune-related mechanism involved in BC patients, we performed immune infiltration analysis using the two BC prognosis models. M2 macrophages were significantly correlated with the BC1 and BC2 models (Figure 4A,4B). Moreover, macrophage M2 infiltration was remarkably higher in the high-risk group than in the low-risk group (Figure 4C,4D).
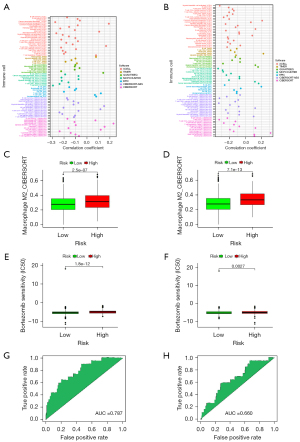
For BC chemotherapy, bortezomib is a first-in-class selective and reversible proteasome inhibitor that targets the 26S proteasome (14). Previous studies have observed that bortezomib combined with antiestrogen therapy might have therapeutic advantages in the management of early-stage BC (15-17). Our study also found that patients in the high-risk group were more sensitive to bortezomib than those in the low-risk group (Figure 4E,4F).
To further test these two models in cohorts from different populations, we assessed the testing power of both models of BC development-related gene sets in GSE4922. The AUC of the BC1 model was 0.79, which was higher than that of the BC2 model, implying that the testing power of the BC1 model was stronger than that of the BC2 model (Figure 4G,4H).
Discussion
Worldwide, especially for women, BC is the main cause of cancer-related death (18). Even with gradually progressing diagnosis and surgical treatments, the recurrence rate and cancer-related death rate of BC are still very high. Hence, it is of great urgency to explore new biomarkers that can precisely diagnose BC and predict prognosis for the treatment and management of BC. In this study, we identified two prognostic models (BC1 and BC2) using bioinformatics analysis on the basis of BC-related DEGs and further explored the prognostic value of BC survival time and the mechanism involved. One model (BC1) was observed to derive a better prognostic risk factor, which classified the survival time in BC patients into low- and high-risk categories.
A number of previous studies have shown that abnormal expression of genes in BC is closely related to disease prognosis and can be used as a potential biomarker of prognosis (19,20). In the present study, a number of genes were differentially expressed in BC at different stages. This indicated that gene expression patterns varied along with BC development. Compared to normal breast tissue, a gene may be differentially expressed in early BC but not in the advanced stage. A total of 738 characteristic BC development genes were identified, including 476 genes that were gradually upregulated and 262 genes that were gradually downregulated with BC development. The development of BC results from interactive effects of multiple genes. Prominently, not all characteristic BC development genes were associated with the prognosis of BC, some of which may result from selection bias or analytical error. Furthermore, DE genes that were upregulated or downregulated may represent entirely different mechanisms involved in BC development. The BC unfavorable gene set comprised 26 gradually upregulated DEGs, and the BC favorable gene set contained 14 gradually downregulated DEGs. Unsurprisingly, previous studies have suggested that some of these genes are associated with BC development (21-23). These results confirmed the possibility that the BC unfavorable gene set and BC favorable gene set could be used as a prognostic model for BC.
In previous studies, a gene coefficient was often obtained from a Cox regression analysis or other method in the training set (24-26). However, due to the limitations of the sample size and the heterogeneity of the tumor, we may never know the true coefficient of a gene. Therefore, GSVA was used to score individual samples against the gene sets (BC unfavorable gene set and BC favorable gene set) in our study. ROC curve analysis suggested that both the BC unfavorable and BC favorable GSVA scores exhibited strong prognostic capacity for BC, which was verified in two other independent datasets. Univariate and multivariate Cox regression analyses suggested that the BC unfavorable and BC favorable gene sets and the risk score systems were independent prognostic factors for BC. This result was also verified in an independent dataset. However, further studies are needed to investigate and validate the functions of these genes, and the synergy between the genes of these two gene sets in BC development still requires molecular experimental validation. Their application in the clinic still needs to wait for further decline in sequencing costs.
In addition, the interaction between the immune microenvironment and cancer cells is important for tumor progression (27). Both the BC development-promoting gene model and BC development inhibitory gene model were associated with high infiltration of M2 macrophages. Both the BC development-promoting gene model and the BC development-suppressing gene model were related to patient drug responsiveness to bortezomib. Naturally, studies aimed at revealing the exact mechanisms and DEGs in BC are advocated. The potential of molecularly targeted immunotherapy to intervene in BC may be promising.
Conclusions
In conclusion, this study explored and validated one prognostic model (BC1) to better diagnose and predict the survival of BC patients using a cluster of 12 DEGs, and extensive functional exploration revealed that the model was closely related to the tumor immune microenvironment, especially M2 macrophage infiltration. Our study provides new insights into further studies in BC, which requires further research.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2444/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2444/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2444/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work and in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Youlden DR, Cramb SM, Dunn NA, et al. The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol 2012;36:237-48. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Ilgun AS, Sarsenov D, Alco G, et al. Long-term survival effects of preoperative breast MRI in patients with breast-conserving surgery. Acta Clin Croat 2022;61:30-7. [Crossref] [PubMed]
- Broeders M, Paci E. The balance sheet of benefits and harms of breast cancer population-based screening in Europe: outcome research, practice and future challenges. Womens Health (Lond) 2015;11:883-90. [Crossref] [PubMed]
- Loibl S, Poortmans P, Morrow M, et al. Breast cancer. Lancet 2021;397:1750-69. [Crossref] [PubMed]
- Xia C, Dong X, Li H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl) 2022;135:584-90. [Crossref] [PubMed]
- Ping Z, Siegal GP, Almeida JS, et al. Mining genome sequencing data to identify the genomic features linked to breast cancer histopathology. J Pathol Inform 2014;5:3. [Crossref] [PubMed]
- Maguire SL, Peck B, Wai PT, et al. Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. J Pathol 2016;240:315-28. [Crossref] [PubMed]
- Zhang G, Wang Y, Chen B, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann Transl Med 2019;7:179. [Crossref] [PubMed]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 2006;7:191. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Lina Y, Ting Q, Tong W. The Application of LASSO in the Cox Model. Chinese Journal of Health Statistics 2012;29:58-60, 64.
- Obuchowski NA, Bullen JA. Receiver operating characteristic (ROC) curves: review of methods with applications in diagnostic medicine. Phys Med Biol 2018;63:07TR01. [Crossref] [PubMed]
- Tan CRC, Abdul-Majeed S, Cael B, et al. Clinical Pharmacokinetics and Pharmacodynamics of Bortezomib. Clin Pharmacokinet 2019;58:157-68. [Crossref] [PubMed]
- Periyasamy-Thandavan S, Jackson WH, Samaddar JS, et al. Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress and induces caspase-dependent cell death in antiestrogen-sensitive and resistant ER+ breast cancer cells. Autophagy 2010;6:19-35. [Crossref] [PubMed]
- Kawaguchi T, Miyazawa K, Moriya S, et al. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol 2011;38:643-54. [PubMed]
- Thaler S, Thiede G, Hengstler JG, et al. The proteasome inhibitor Bortezomib (Velcade) as potential inhibitor of estrogen receptor-positive breast cancer. Int J Cancer 2015;137:686-97. [Crossref] [PubMed]
- Ahmad A. Breast Cancer Statistics: Recent Trends. Adv Exp Med Biol 2019;1152:1-7. [Crossref] [PubMed]
- Joe S, Nam H. Prognostic factor analysis for breast cancer using gene expression profiles. BMC Med Inform Decis Mak 2016;16:56. [Crossref] [PubMed]
- Mranda GM, Xue Y, Zhou XG, et al. Revisiting the 8th AJCC system for gastric cancer: A review on validations, nomograms, lymph nodes impact, and proposed modifications. Ann Med Surg (Lond) 2022;75:103411. [Crossref] [PubMed]
- Singh Y, Subbarao N, Jaimini A, et al. Genome-wide expression reveals potential biomarkers in breast cancer bone metastasis. J Integr Bioinform 2022;19:20210041. [Crossref] [PubMed]
- Du Y, Han Y, Wang X, et al. Identification of Immune-Related Breast Cancer Chemotherapy Resistance Genes via Bioinformatics Approaches. Front Oncol 2022;12:772723. [Crossref] [PubMed]
- Hou L, Hou S, Yin L, et al. Epithelial-Mesenchymal Transition-Based Gene Signature and Distinct Molecular Subtypes for Predicting Clinical Outcomes in Breast Cancer. Int J Gen Med 2022;15:3497-515. [Crossref] [PubMed]
- Hao S, Huang M, Xu X, et al. MDN1 Mutation Is Associated With High Tumor Mutation Burden and Unfavorable Prognosis in Breast Cancer. Front Genet 2022;13:857836. [Crossref] [PubMed]
- Wang J, Zhang X, Li J, et al. ADRB1 was identified as a potential biomarker for breast cancer by the co-analysis of tumor mutational burden and immune infiltration. Aging (Albany NY) 2020;13:351-63. [Crossref] [PubMed]
- Zhang R, Li Q, Fu J, et al. Comprehensive analysis of genomic mutation signature and tumor mutation burden for prognosis of intrahepatic cholangiocarcinoma. BMC Cancer 2021;21:112. [Crossref] [PubMed]
- Ni Y, Tsang JY, Shao Y, et al. Combining Analysis of Tumor-infiltrating Lymphocytes (TIL) and PD-L1 Refined the Prognostication of Breast Cancer Subtypes. Oncologist 2022;27:e313-27. [Crossref] [PubMed]


