
Comprehensive pan-cancer analysis identifies FHL2 associated with poor prognosis in lung adenocarcinoma
Highlight box
Key findings
• FHL2 plays an important role in tumorigenesis and tumor progression in multiple cancers, which was significantly associated with the tumor microenvironment and may be involved in post-transcriptional modification
What is known and what is new?
• FHL2 promotes the progression of ovarian, cervical and pancreatic cancers
• Upregulated FHL2 was associated with poor prognosis in a variety of cancers and was significantly associated with fibroblast infiltration and tumor stage, particularly in lung adenocarcinoma
What is the implication, and what should change now?
• FHL2 was association with poor prognosis and provide guidance for future mechanistic exploration. Such as EMT and immune cell infiltration.
Introduction
Cancer is one of the world’s leading public health problems, causing a large number of deaths each year (1). Lung cancer is the most frequently diagnosed cancer in 36 countries, accounting for 18% of cancer deaths, and it comprises a variety of histological subtypes, of which lung adenocarcinoma (LUAD) is the most common type (2). Despite the great advances in therapeutic methods for lung cancer, the prognosis of some patients remains poor and there is an urgent requirement to identify new therapeutic targets and biomarkers in LUAD (3,4).
Cellular processes are dependent on the synergy of multiple proteins (5). Four-and-a-half-LIM-only protein (FHL) family members are important mediators of protein-protein interaction and exhibit specificity in tissue development and organ expression patterns. The FHL family contains five multifunctional proteins (FHL1-5) that are involved in cell survival, transcriptional regulation, and signal transduction (6). Among these proteins, FHL2 is one of the most reported members in tumors. As its name shows, FHL2 has a half-LIM amino-terminal domain followed by four complete LIM domains (7). Interestingly, LIM domains cannot bind directly to DNA, but rather act as specific scaffold structures that bind to various proteins to perform different functions (8). Studies have shown that aberrant expression of FHL2 is closely associated with the progression of multiple cancers where it regulates many key cellular processes (cell proliferation, migration and apoptosis). For example, FHL2 promotes MDM2-mediated degradation of IER3 to regulate the growth of cervical cancer cells (9). miR-340 targets FHL2 to inhibit the Wnt/β-catenin pathway in ovarian cancer (10). Decreased expression of FHL2 led to a significant decrease in cell survival, proliferation and radiation resistance in pancreatic cancer (11). Overall, FHL2 plays an essential role in a variety of tumors and deserves further investigation.
Based on the current state of research and the rapid development of high-throughput sequencing technology, we can further explore the potential value of FHL2 in different cancers in a comprehensive and in-depth manner using bioinformatics approaches. In this study, we explored the expression level and prognostic significance of FHL2 using R packages, and mined the correlation between FHL2 and immune infiltration levels in tumor tissues. In addition, multiple functional enrichment algorithms were used to reveal that FHL2 may influence the progression of LUAD by regulating epithelial mesenchymal transition. The findings of this study are expected to further guide more in-depth molecular mechanism research. We present this article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2786/rc).
Methods
Gene expression analysis
We used TIMER2.0 online tool (http://timer.comp-genomics.org/) to compare the mRNA expression of FHL2 between tumor and adjacent normal tissues from The Cancer Genome Atlas (TCGA) and across 36 types of cancers (12,13). We also downloaded RNAseq data in TPM (transcripts per million reads) format after log2 (x+1) conversion from the Xena database (https://xenabrowser.net/datapages/), selecting paired samples to study the expression of FHL2 in 18 cancers. Besides, GSE31210 and GSE10072 were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Survival and clinical analysis
The samples were divided into high- and low-expression groups based on the median expression of FHL2 derived from the gene expression profile data. With the Kaplan-Meier method, we used the R packages “survival” and “survminer” to analyze the survival information and draw the survival curves. In addition, the “forest plot” package was used to visualize the results of the Cox analysis for survival data (14).
Distribution of FHL2 expression in molecular subtypes and immune subtypes of fourteen types of human tumors
TISIDB (http://cis.hku.hk/TISIDB/) is a website for tumor and immune interaction, integrating various heterogeneous data types (15). We investigated the correlation between FHL2 expression and the molecular subtypes of tumors as well as immune subtypes in pan-cancer based on the TISIDB database. Molecular subtypes are different depending on the tumors, and immune subtypes include C1 (wound healing); C2 (IFN-gamma dominant); C3 (inflammatory); C4 (lymphocyte depleted); C5 (immunologically quiet); C6 (TGF-β dominant).
Immune infiltration analysis
Immune infiltration analysis can visualise the correlation between the expression of FHL2 and the abundance of cells in the immune microenvironment. In the present study, we use the R package “IOBR” (version 0.99.9) by the means of “deconvo_EPIC” to evaluate the B cells, CAFs, CD4_Tcells, CD8_Tcells, Endothelial, Macrophages, NK cells infiltration scores for each patient and each tumor (16,17). To further confirm the results obtained, we used the TIMER 2.0 database to perform a more in-depth analysis of FHL2 and cancer-associated fibroblasts. Ultimately, we incorporated three algorithms (EPIC, MCPCOUNTER, TIDE).
Enrichment analysis
The R package “limma”(version 3.40.6) was used to analyze differentially expressed genes between different expression groups of FHL2 (18). Enrichment analysis of Gene Ontology and Kyoto Encyclopedia of Genes and Genomes were performed using the R package “Cluster Profiler” (version 3.14.3). Kyoto Encyclopedia of Genes and Genomes (KEGG) (c2.cp.kegg.v7.4.symbols.gmt) and Hallmark (h.all.v7.4.symbols.gmt) gene sets were downloaded from the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp). The samples were divided into two subgroups based on the expression of FHL2 to explore relevant pathways and molecular mechanisms (19).
Genetic mutation analysis
cBioPortal (https://www.cbioportal.org/), a strong online tool, was used to analyze the mutation of FHL2. In addition, the analysis of somatic mutations in LUAD samples was also downloaded from the TCGA database website with the format of “maf”, using the R package “Maftools” to draw the waterfall mapping, which can help visualize the results of the analysis of mutant genes (20,21).
CancerSEA database
We use the CancerSEA website to process single-cell analysis. The data were collected from cancer-related scRNA-seq datasets in human from Sequence Read Archive (SRA), GEO and ArrayExpress. Functional analysis was performed from data sets such as HCMDB, Cyclebase and StemMapper, redefining a total of 14 functional states.
Statistical analysis
R software (V.3.6.1) was utilized for data analysis. The significance between the two groups was identified using Wilcox test. For statistical analysis of non-normally distributed variables, the Wilcoxon test was applied for differences between two groups, and the Kruskal Wallis test was applied for differences between three or more groups. The Spearman algorithm was used for the correlation analysis. The survival time differences between the two risk groups were estimated using Kaplan-Meier curves and log-rank test.
Results
Expression pattern of FHL2 across cancers
To investigate mRNA expression levels of FHL2 in a wide range of cancers, we used the Tumor Immune Estimation Resource (TIMER2.0) database. “Diff Exp” module was applied to explore differential gene expression between tumor and normal tissues (Figure 1A). FHL2 was found to be highly expressed in cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), Head and Neck squamous cell carcinoma (HNSC), LUAD, lung squamous cell carcinoma (LUSC), while down-expressed in breast invasive carcinoma (BRCA), Kidney Chromophobe (KICH), Liver hepatocellular carcinoma (LIHC), prostate adenocarcinoma (PRAD), thyroid carcinoma (THCA), Uterine Corpus Endometrial Carcinoma (UCEC). In addition, to gain a more complete understanding of FHL2 mRNA expression levels in cancers, we downloaded TCGA RNA-seq data in TPM format from the UCSC XENA. By comparing the expression of FHL2 in cancer tissues with matched normal tissues, we also found that FHL2 is up-regulated in COAD, HNSC, LUAD, LUSC, while down-regulated in BRCA, KICH, LIHC, PRAD, and THCA (Figure 1B,1C). The significant heterogeneity of tumors, with huge differences in disease progression, radio-chemotherapy sensitivity and prognosis between patients, the shift from traditional morphological typing to molecular typing is more beneficial for accurate diagnosis of cancer. Pre-calculated correlations between genes and immune functions (e.g., lymphocytes, immunomodulatory molecules and chemokines), TISIDB Tumor Immune System Interactions and Drug Bank (TISIDB) database classifies a wide range of tumors into molecular subtypes. By using TISIDB database, we found that based on FHL2, patients can be classified into multiple molecular subtypes and that FHL2 expression varies among molecular subtypes (P<0.05) (Figure 2), which implied that the expression of FHL2 is related to patient prognosis.


Prognostic value of FHL2 in multiple malignancies
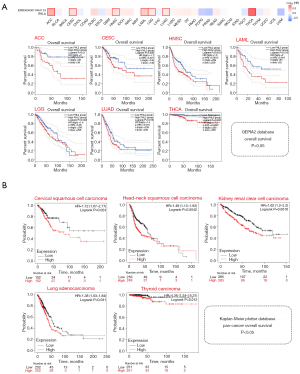
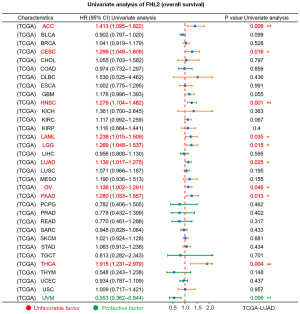
We explored the prognostic significance of FHL2 in multiple cancers using the GEPIA2 database (Figure 3A). Results showed that FHL2 expression in adrenocortical carcinoma (ACC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), HNSC, Acute Myeloid Leukemia (LAML), Brain Lower Grade Glioma (LGG), LUAD, and THCA was associated with overall survival. In this group of 7 cancers, patients with high FHL2 expression had a worse prognosis. Further, we analyzed the relationship between FHL2 expression and overall survival using a pan-cancer analysis module in the Kaplan-Meier Plotter database (Figure 3B). Results revealed that patients with elevated FHL2 expression in CESC, HNSC, KIRC, LUAD, and THCA had worse patient outcomes. To more fully investigate the prognostic value of FHL2, we also performed a univariate Cox analysis in cancers (TCGA database). We identified FHL2 as an adverse prognostic factor in ACC, CESC, HNSC, LAML, LGG, LUAD, ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), THCA, and a protective factor in uveal melanoma (UVM) (Figure 4). These results suggested that FHL2 may plays a critical role in overall survival of cancer patients.
Analysis of tumor immune microenvironment and mRNA modification across cancers
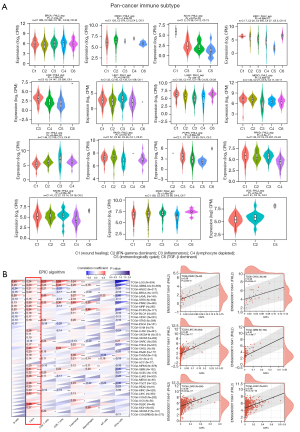
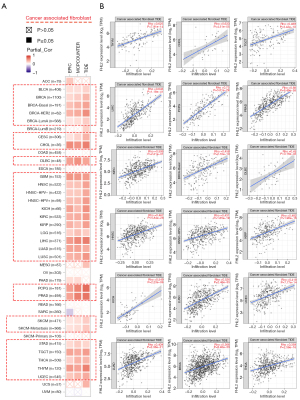
The tumor immune microenvironment plays an important role in the development of tumors. As with the molecular subtypes, using the TISIDB database, we found that patients could be classified into six immune subtypes based on the association of FHL2 expression with immune function (Figure 5A). They are: C1 (wound healing); C2 (IFN-gamma dominant); C3 (inflammatory); C4 (lymphocyte depleted); C5 (immunologically quiet); C6 (TGF-β dominant). Then we performed a pan-cancer analysis of eight immune-related cell types using the EPIC algorithm and found that FHL2 was significantly associated with cancer associated fibroblasts (CAFs) in a wide range of cancers (Figure 5B). Further, to investigate the correlation between FHL2 and CAFs, we used the TIMER 2.0 database to obtain an immune landscape of FHL2 and CAFs in pan-cancer based on three algorithms. As shown in Figure 6A, mRNA expression of FHL2 is highly correlated with CAFs infiltration levels and we exhibited scatter plots calculated by the TIDE algorithm in 18 cancers (Figure 6B). In addition, we investigated the relationship between FHL2 and mRNA modification genes (m1A, m5C, m6A). The results showed that the mRNA level expression of FHL2 correlated significantly with that of mRNA modified genes in most cancer types, except for UVM and GBM (Figure S1A). Since immunomodulators are closely associated with tumor escape, we investigated the association of immunoinhibitors, immunostimulators, and MHC molecules with FHL2 in pan-cancer, enabling targeted selection of immunotherapies (Figure S1B). Taken together, these studies indicated that FHL2 may be associated with tumor immune cell infiltration and genes of post-transcriptional modification.
Expression pattern and clinical relevance of FHL2 in LUAD
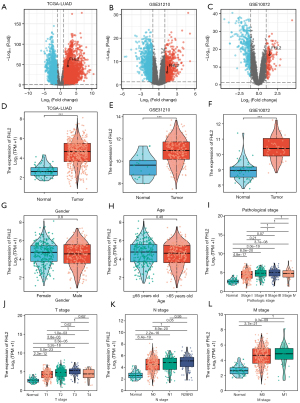
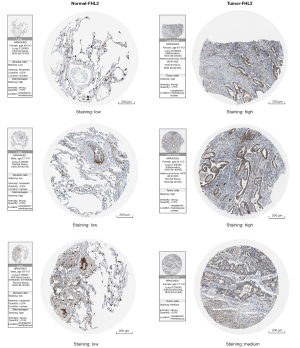
Combining TCGA, GSE31210, and GSE10072 expression profiles, we further validated high FHL2 expression in LUAD (Figure 7A-7F). The correlation between FHL2 expression and clinicopathologic factors was also analyzed. We found no statistically significant difference in FHL2 expression by gender or age (Figure 7G,7H). FHL2 was significantly differentially expressed between T1 stage and T2, T3 stage, in addition to lymph node metastasis and absence of metastasis. (Figure 7I-7L). Using the Human Protein Atlas (HPA) database, we found high expression of FHL2 at the protein level in tumors (Figure 8). Further, we validated its prognostic value in the TCGA and GSE31210 databases. The results indicated that high FHL2 expression has a poor prognosis in both overall survival, disease-free survival, and progression-free survival. Univariate and multivariate Cox analysis of GSE31210 revealed FHL2 was an independent prognostic factor (Figure S2).
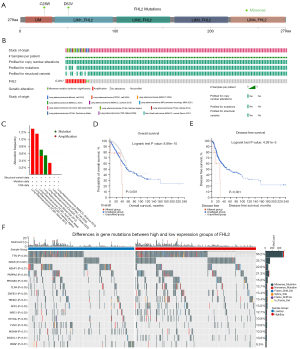
Relationship between FHL2 and mutations
First, we used cBioPortal to identify somatic mutations in FHL2. We found that FHL2 has two loci of mutation in the form of Missense (Figure 9A). In addition, we selected 12 databases of LUAD. The overall probability of FHL2 mutation was found to be 0.8% in the samples (Figure 9B). The mutations mainly take the form of amplification (Figure 9C). By comparing the prognosis of the mutant and non-mutant groups, we found that the mutant group had a worse prognosis in terms of overall survival and disease-free survival (Figure 9D,9E). Further, we performed mutation landscape analysis of the TCGA-LUAD database based on FHL2 expression. As shown in the diagram, Figure 9F displayed the 15 genes with differential mutation frequencies in the FHL2 high and low expression groups. These results suggested expression of FHL2 in lung cancer significantly correlates with gene mutation rate.
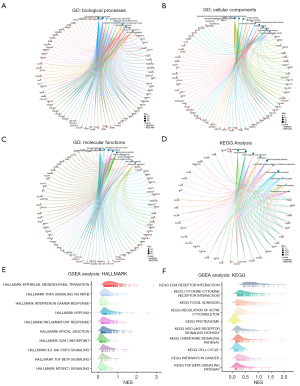
Significant correlation between FHL2 and EMT
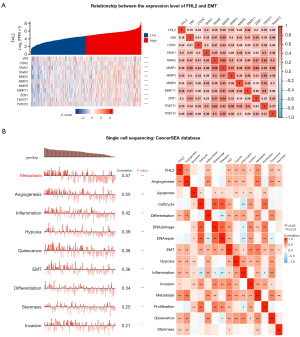
To further explore the potential role of FHL2 in LUAD. We performed GO, KEGG enrichment analysis of the top 200 FHL2-associated differentially expressed genes (FDR <0.05, |log2FC| >1). We found a strong correlation between FHL2 and cellular adhesion, ECM-receptor interaction, and PI3K-AKT signaling pathways (Figure 10A-10D). To further validate these results, we selected HALLMARK and KEGG gene sets for GSEA enrichment analysis of FHL2 (Figure 10E,10F). We found that FHL2 significantly enriched EMT-associated pathways, cell cycle, and other pathways associated with tumor progression. In addition, we analyzed the association of FHL2 with EMT-associated markers in the TCGA-LUAD database. Results showed that FHL2 was associated with VIM (vimentin), CDH2 (Cadherin 2), SNAI1 (Snail Family Transcriptional Repressor 1), SNAI2 (Snail Family Transcriptional Repressor 2), MMP2 (Matrix Metallopeptidase 2), MMP3 (Matrix Metallopeptidase 3), MMP9 (Matrix Metallopeptidase 9), MMP11 (Matrix Metallopeptidase 11), ZEB1 (Zinc Finger E-Box Binding Homeobox 1), TWIST1 (Twist Family BHLH Transcription Factor 1), and TWIST2 (Twist Family BHLH Transcription Factor 2) (Figure 11A). These results were also validated by a single-cell sequencing database. By using LUAD cell data from the Cancer SEA database, we found that FHL2 was associated with metastasis, EMT, and invasion at the single-cell level (Figure 11B). We reasonably hypothesize that FHL2 expression is involved in the EMT process in LUAD and can influence the expression of markers of EMT.
Discussion
According to the current study, there is no study on the potential prognostic impact and biological function of FHL2 in pan-cancer. In this study, we combined the UCSC XENA and TISIDB databases to analyze the mRNA expression of FHL2, and found that the expression of FHL2 varies significantly between different cancers. These results consistent with previous studies, such as those on colon cancer (22) and kidney chromophobe (23). Next, the relationship between FHL2 expression and prognosis was analyzed using GEPIA2, Kaplan Meier plotter, and UCSC databases. These results further confirmed that FHL2 might act as the biomarker of these cancers. To sum up, FHL2 may play different roles in various cancers.
Different molecular and immune subtypes of tumors have distinct degrees of progression. Bioinformatics analysis showed that the expression of FHL2 correlated with molecular and immune subtypes in a variety of tumors. In-depth exploration of these correlations could help to uncover the potential mechanisms of FHL2. In addition, the significant correlation between FHL2 expression and chemokines and immunomodulatory factors in many cancers also indicates that FHL2 may play a role in regulating immunomodulatory factor expression.
Interestingly, FHL2 shows a very consistent positive relationship with CAFs in pan cancers. CAFs, located around cancer tissues, are a heterogeneous population of irreversibly activated fibroblasts with distinct functions (24). Nowadays, more and more studies have shown that CAFs can promote or inhibit the process of tumor by affecting the beginning, process, immune escape, and metastasis of tumor (24-28). We analyzed the relationship between the expression of FHL2 and the degree of infiltration in pan cancers. It is highly demonstrated that positive relationships, indicated that FHL2 plays an important role in the infiltration of CAFs. Previous study has shown that FHL2 has been associated with fibroblast formation in normal cells (29). The most classical representation is that FHL2 is intensely upregulated during the skin injury, presenting the highest expression in the migration and proliferation phase and decreasing once again in the later stages (30). Meanwhile, in the wounded area, by the means of promoting migration, FHL2 promotes integrated fibroblasts to replace blood clots, form granulation tissue and ensure its contraction, which is essential for wound edge healing (29,31). As for the way of the mechanism, previous study indicated that TGF-β positively stimulates FHL2 transcription, and FHL2 protein supports TGF-β-driven EMT progression (32). The study about the co-localization of FHL2 and α-SMA implies TGF-β Induced FHL2 may be secreted into CAFs (32), which coincides with our relationship between FHL2 and CAFs.
mRNA modification, as a novel and essential modification form in the field of epigenetics, acts as a new way of post-transcriptional gene regulation (33). RNA modifications play an important role in the regulation of gene expression, tumor development, invasion and metastasis. RNA modifications are dynamic, reversible and widespread (34). There are currently more than 170 known chemical modifications of RNA, and methylation is one of the most important RNA modifications, including N1-methyladenosine (m1A), 5-methylcytosine (m5C), N6-Methyladenosine (m6A). m6A has been most widely reported, which is related to mRNA stability, splicing processing, translation (35). Therefore, exploring the relationship between FHL2 and mRNA modification in pan-cancer may be another direction to study the function and mechanism of FHL2. The analysis of the association of mRNA modification genes with FHL2 in pan-cancer analysis indicates that FHL2 plays a positive role in most cancers but not in UVM and GBM.
Next, we combined the results of pan-cancer analysis and found that the expression of FHL2 in LUAD was high in all databases, while the results of various prognostic analyses were poor. Meanwhile, there was a positive correlation with CAFs, indicating that FHL2 may play a crucial role in LUAD. However, most previous published studies in lung cancer only focused on the gene expression profiles or just on the EMT markers (36), which had limitations to understand the mechanisms and characteristics of FHL2. Therefore, there is a pressing need to uncover potential therapeutic targets using multi-layer data analysis.
Here, a comprehensive bioinformatics analysis was performed via using RNA-seq data and prognostic information from various databases including TCGA and GEO, indicating that the upregulated FHL2 expression is associated with poor prognosis, suggesting that FHL2 may be used as a risk factor to predict overall survival in LUAD. Besides, our results also showed that FHL2 mutated more in the high expression of FHL2. Previous study has shown that the occurrence of cancer is due to the accumulation of somatic mutations and other genetic alternations which damage the cell proliferation and eventually tumorigenesis (37). The positive correlation between the FHL2 expression and mutation rate may be an important factor in promoting cancer progression. Here, we found that mutated FHL2 was associated with poor prognosis in LUAD, the mutation promoted the function of FHL2, which is also a complement to the role of FHL2.
GSEA and KEGG enrichment analysis showed that targeting the EMT relative pathway such as NF-κB and TGF-β might be an important mechanism for the effect of FHL2. Therefore, we further explore the relationship between FHL2 and EMT relative genes, indicating that FHL2 showed positive correlation with most EMT-related genes, while the single-cell sequencing results further predicted that FHL2 may be involved in lung cancer metastasis, angiogenesis, inflammation and hypoxia processes, which is also consistent with the results of GSEA enrichment analysis, but still needs to be demonstrated by future in-depth experiments. Overall, the results of our bioinformatics analysis suggest that FHL2 expression may have the potential to indicate the prognosis of a variety of cancers such as LUAD.
Conclusions
Upregulated FHL2 was associated with poor patient prognosis in a variety of cancers and was significantly associated with fibroblast infiltration and tumor stage, particularly in LUAD. Meanwhile, FHL2 expression was significantly correlated with the expression of mRNA-modified genes in a variety of cancers, and it was observed that FHL2 expression may influence the expression levels of chemokines as well as immunomodulators. In addition, the functional study of FHL2 indicated that FHL2 may involve in EMT, especially through TGF-β signal pathway in LUAD. Prospective and experimental studies of FHL2 expression and immune cell infiltration in different cancer populations in the future may provide additional insights into tumor mechanisms to guide strategies for treatment.
Acknowledgments
Funding: This work was supported by the grants from
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2786/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2786/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-2786/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Huang B, Deng W, Chen P, et al. Development and validation of a novel ubiquitination-related gene prognostic signature based on tumor microenvironment for colon cancer. Transl Cancer Res 2022;11:3724-40. [Crossref] [PubMed]
- Johannessen M, Møller S, Hansen T, et al. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci 2006;63:268-84. [Crossref] [PubMed]
- Zhou R, Li S, Liu J, et al. Up-regulated FHL2 inhibits ovulation through interacting with androgen receptor and ERK1/2 in polycystic ovary syndrome. EBioMedicine 2020;52:102635. [Crossref] [PubMed]
- Kurakula K, Sommer D, Sokolovic M, et al. LIM-only protein FHL2 is a positive regulator of liver X receptors in smooth muscle cells involved in lipid homeostasis. Mol Cell Biol 2015;35:52-62. [Crossref] [PubMed]
- Cao CY, Mok SW, Cheng VW, et al. The FHL2 regulation in the transcriptional circuitry of human cancers. Gene 2015;572:1-7. [Crossref] [PubMed]
- Jin H, Lee K, Kim YH, et al. Scaffold protein FHL2 facilitates MDM2-mediated degradation of IER3 to regulate proliferation of cervical cancer cells. Oncogene 2016;35:5106-18. [Crossref] [PubMed]
- Huang Z, Li Q, Luo K, et al. miR-340-FHL2 axis inhibits cell growth and metastasis in ovarian cancer. Cell Death Dis 2019;10:372. [Crossref] [PubMed]
- Zienert E, Eke I, Aust D, et al. LIM-only protein FHL2 critically determines survival and radioresistance of pancreatic cancer cells. Cancer Lett 2015;364:17-24. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 2017;35:314-6. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-416.e11. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Zeng D, Ye Z, Shen R, et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front Immunol 2021;12:687975. [Crossref] [PubMed]
- Racle J, de Jonge K, Baumgaertner P, et al. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017;6:e26476. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Liu Y, He Q, Sun W. Association analysis using somatic mutations. PLoS Genet 2018;14:e1007746. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Wang J, Yang Y, Xia HH, et al. Suppression of FHL2 expression induces cell differentiation and inhibits gastric and colon carcinogenesis. Gastroenterology 2007;132:1066-76. [Crossref] [PubMed]
- Xu J, Zhou J, Li MS, et al. Transcriptional regulation of the tumor suppressor FHL2 by p53 in human kidney and liver cells. PLoS One 2014;9:e99359. [Crossref] [PubMed]
- Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016;16:582-98. [Crossref] [PubMed]
- Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174-86. [Crossref] [PubMed]
- Bussard KM, Mutkus L, Stumpf K, et al. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res 2016;18:84. [Crossref] [PubMed]
- Steven A, Seliger B. The Role of Immune Escape and Immune Cell Infiltration in Breast Cancer. Breast Care (Basel) 2018;13:16-21. [Crossref] [PubMed]
- Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18:99-115. [Crossref] [PubMed]
- Wixler V, Hirner S, Müller JM, et al. Deficiency in the LIM-only protein Fhl2 impairs skin wound healing. J Cell Biol 2007;177:163-72. [Crossref] [PubMed]
- Wixler V. The role of FHL2 in wound healing and inflammation. FASEB J 2019;33:7799-809. [Crossref] [PubMed]
- Gullotti L, Czerwitzki J, Kirfel J, et al. FHL2 expression in peritumoural fibroblasts correlates with lymphatic metastasis in sporadic but not in HNPCC-associated colon cancer. Lab Invest 2011;91:1695-705. [Crossref] [PubMed]
- Zhang W, Jiang B, Guo Z, et al. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis 2010;31:1220-9. [Crossref] [PubMed]
- Liang W, Lin Z, Du C, et al. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer 2020;19:38. [Crossref] [PubMed]
- Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer 2020;20:303-22. [Crossref] [PubMed]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31-42. [Crossref] [PubMed]
- Li N, Xu L, Zhang J, et al. High level of FHL2 exacerbates the outcome of non-small cell lung cancer (NSCLC) patients and the malignant phenotype in NSCLC cells. Int J Exp Pathol 2022;103:90-101. [Crossref] [PubMed]
- Iranzo J, Martincorena I, Koonin EV. Cancer-mutation network and the number and specificity of driver mutations. Proc Natl Acad Sci USA 2018;115: [Internet]. [Crossref] [PubMed]


