
A novel genes-based signature with prognostic value and predictive ability to select patients responsive to Atezolizumab treatment in bladder cancer: an analysis on data from real-world studies
Highlight box
Key findings
• We proposed two molecular mRNAs that can help screen patients with advanced bladder cancer suitable for immune checkpoint inhibitors.
What is known and what is new?
• Although immune checkpoint inhibitors are currently approved for clinical use in the treatment of advanced bladder cancer, the selection of patients responsive to immune checkpoint inhibitor therapy remains a major clinical challenge.
• In this study, we report that CXCL9/10 mRNA can be used as molecular markers to predict response to Atezolizumab treatment, with a potential prognostic significance in bladder cancer patients.
What is the implication, and what should change now?
• The joint application of multiple molecular markers would be the ideal approach for screening patients who are suitable for immune checkpoint inhibitors.
Introduction
The use of immune checkpoint inhibitors (ICIs) has received widespread attention as tumor therapy has entered the era of immunotherapy. Clinical study has shown promising data regarding the use of immunotherapeutic strategies in bladder cancer treatment (1). Though these therapies can improve the disease prognosis, some patients exhibit low or no response due to high tumor heterogeneity. The therapy’s high toxic effects and the oncological disadvantage of selecting the wrong treatment for unresponsive patients underscore the importance of identifying biomarkers for treatment response. A recent study has demonstrated the possibility of utilizing upfront biomarkers of resistance to ICI therapy in bladder cancer (2). Currently, prediction of treatment response mainly relies on programmed cell death 1 ligand 1 (PD-L1) expression and tumor mutation burden (TMB) values (3,4). However, some patients with low PD-L1 expression can still benefit from treatment with ICIs, and TMB has the disadvantage of being a time-consuming and expensive test.
In urological tumors, mRNA has been shown to be a potentially useful biomarker, but the large amount of available research data is yet to be implemented in clinical practice. Nonetheless, it is clear that different cancers exhibit varying clinical behaviors in different patients, making the use of biomarkers a promising strategy. Epigenetic-based biomarkers, including aberrant DNA methylations, dysregulated expression of chromatin structure proteins, and miRNAs, nuclear transfer RNA (nt-RNAs), or long non-coding RNAs (lncRNAs), could have a significant impact on clinical practice in oncology (5). Advanced methodologies and technologies have enabled molecular subtyping of bladder cancer cases, offering clues for selecting appropriate therapies upfront, before beginning treatment. Although a recent paper demonstrated the possibility of patient-specific immune profiling of bladder cancer cases to identify which patients will respond to immunotherapy (2). However, no specific mRNA biomarkers are currently in use for clinical practice. In this study, we used data from real-world clinical studies to help us screen for effective molecular markers. Two datasets, IMvigor210 (6) and GSE176307 (7), contain clinical information and treatment responses of two groups of patients with bladder cancer treated with ICIs, from which we analyzed the gene expression of two groups of patients who were effective and ineffective for treatment with the PD-L1 inhibitor Atezolizumab, and screened the differentially expressed genes as molecular markers to predict treatment response. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-220/rc).
Methods
Data sources
The genetic sequencing results and clinical information of bladder cancer patients treated with Atezolizumab in the IMvigor210 (4) and GSE176307 (5) datasets were selected and divided into response and non-response groups according to treatment outcome of partial remission (PR), complete remission (CR), or disease progression (PD) and disease stabilization (SD). In the IMvigor210 dataset, a total of 348 patients were treated with Atezolizumab, 68 treatment-responsive, 230 non-response, and 50 patients whose outcomes were not evaluated were not included in this study. A total of 35 patients received Atezolizumab, 9 treatment-responsive, and 26 non-response in the GSE176307 dataset. The bladder cancer tissue RNA sequencing (RNAseq) data used to analyze correlations between gene and TMB expression were obtained from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), and a total of 401 bladder cancer tissues were included in the analysis by removing incomplete clinical information and normal tissues. Log2 conversion was performed on the RNAseq data in TPM format. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Bioinformatics analysis
R software (version 4.2.1, for statistical analysis and visualization) was used for bioinformatics analysis. The Limma package (version 3.40.2) of R software was used to study the differential expression of mRNA. Gene expression correlation and correlation between gene expression and TMB levels were performed by the R package ggplot2 package (version 3.3.6). pROC package (version 1.17.0.1) and ggplot2 package (version 3.3.6) were used to construct the receiver operating characteristic (ROC) curve. The Survminer package (version 0.4.9) and the surv package (version 3.3.1) were used for Kaplan-Meier (KM) curve plotting.
Statistical analysis
Differences in gene expression between groups were analyzed using the Mann-Whitney U test. Spearman’s correlation coefficient was used to respond to gene expression. Log-rank test was used for prognostic analysis of KM curves. The high and low gene expression groups were divided using median. P<0.05 was considered statistically significant and multiplicity >2 was considered differentially expressed.
Results
Screening for differentially expressed genes
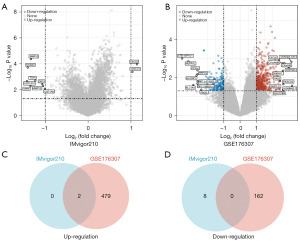
A total of 10 differentially expressed genes were identified in the IMvigor210 dataset for patients responding to Atezolizumab treatment compared to non-responding patients, of which 2 genes were upregulated [P<0.05, log|fold change (FC)| >1] and 8 genes were downregulated (P<0.05, log|FC| >1) (Figure 1A). In GSE176307 dataset of patients responding to Atezolizumab treatment compared to non-responding patients, 643 differentially expressed genes were identified, of which 481 genes were upregulated (P<0.05, log|FC| >1) and 162 genes were downregulated (P<0.05, log|FC| >1) (Figure 1B). Two differentially expressed genes, CXC motif chemokine ligand 9 (CXCL9) and CXC motif chemokine ligand 10 (CXCL10), were co-upregulated in the two datasets (Figure 1C). No differentially expressed genes were co-downregulated in the two datasets (Figure 1D).
Expression and correlation of CXCL9/10 genes and PD-L1 gene/TMB in bladder cancer patients treated with Atezolizumab
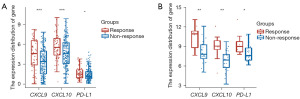
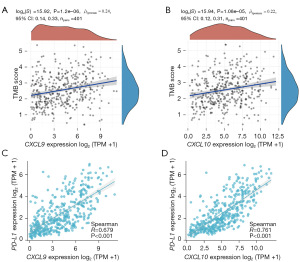
In the IMvigor210 dataset, CXCL9/10 expression was higher in the Atezolizumab treatment-response group than in the non-response group (both P<0.001), and PD-L1 expression did not differ between the two groups (Figure 2A). In the GSE176307 dataset, CXCL9/10 mRNA expression was higher in the Atezolizumab treatment-response group than in the non-response group (both P<0.01), and PD-L1 expression was higher in the Atezolizumab treatment-response group (P<0.05) (Figure 2B). CXCL9/10 mRNA expression was positively correlated with TMB score in bladder cancer tissues in the TCGA database (both P<0.05) (Figure 3A,3B). CXCL9/10 mRNA expression was also positively correlated with PD-L1 expression (P<0.001) (Figure 3C,3D).
Analysis of the predictive efficacy of CXCL9/10 and PD-L1 genes expression on the therapeutic effect of Atezolizumab
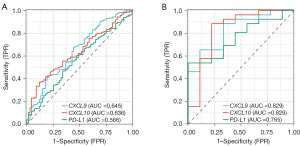
In the IMvigor210 dataset, the area under the ROC curve (AUC) for CXCL9, CXCL10 and PD-L1 mRNA expression predicting response to treatment with Atezolizumab were 0.645, 0.636 and 0.566, respectively (Figure 4A). In the GSE176307 dataset, the AUC for CXCL9, CXCL10 and PD-L1 mRNA expression predicting response to treatment with Atezolizumab were 0.829, 0.829 and 0.765, respectively (Figure 4B).
CXCL9/10 genes expression and prognosis in bladder cancer patients treated with Atezolizumab
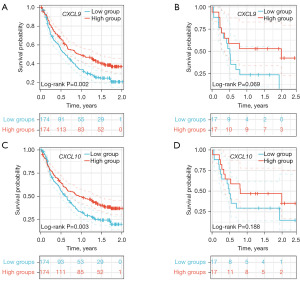
In the IMvigor210 dataset, CXCL9 mRNA was effective in predicting overall survival (OS) in patients treated with Atezolizumab (P=0.002) (Figure 5A), but in the GSE176307 dataset, CXCL9 mRNA was not effective in predicting OS in patients treated with Atezolizumab (P=0.069) (Figure 5B). In the IMvigor210 dataset, CXCL10 mRNA was effective in predicting OS in patients treated with Atezolizumab (P=0.003) (Figure 5C), but in the GSE176307 dataset, CXCL10 was not effective in predicting OS in patients treated with Atezolizumab (P=0.188) (Figure 5D).
Discussion
Although ICIs are currently approved for clinical use in the treatment of advanced bladder cancer, the selection of patients who will respond to ICI therapy remains a major clinical challenge, with approximately 1/3 of patients benefiting from immunotherapy (8). Therefore, the development of molecular markers for determining whether patients respond to ICI therapy is one of the main research directions at present. The findings of a recent study showed that utilizing biomarkers upfront in bladder cancer could predict resistance to ICI therapy (5). Some current studies suggest that molecular markers such as PD-L1, TMB, microsatellite instability, and CD8+ T cells can help guide us to select a superior population suitable for ICI therapy and can improve patient outcomes, especially PD-L1 and TMB are currently compared as molecular markers for predicting the effect of immunotherapy, including bladder cancer (9-11). While biomarkers such as PD-L1 detection and TMB have shown potential benefits in predicting treatment efficacy, they also have possible drawbacks. For instance, an over-reliance on PD-L1 detection can lead to some patients being missed for treatment even if they respond to it but have low PD-L1 expression (12). Additionally, the clinical prediction threshold and value of TMB are inconsistent across different tumors, as it is better in some cancers like melanoma and non-small cell lung cancer (13,14), but not all solid tumors (15). Furthermore, our analysis found that PD-L1 mRNA expression was not significantly different between the Atezolizumab treatment-response and non-response groups, indicating that it may not serve well as a predictor of treatment efficacy. By differential expression analysis, CXCL9/10 mRNA expression was higher in both IMvigor210 and GSE176307 datasets in the responding group than in the non-responding group, and both had higher AUC values than PD-L1 for predicting treatment efficacy, predicting that they may have better predictive efficacy. CXCL9/10 are both small molecular weight cytokines belonging to the CXC chemokine family, which can be produced by antigen-presenting cells (dendritic cells or macrophages) and tumor cells in the tumor microenvironment and lead to the recruitment of C-X-C motif chemokine receptor 3 (CXCR3)-expressing immune cells to the tumor to participate in the immune response (16). Hoch et al. performed a spatial histological analysis of immune cells and chemokines from 69 patients with stage III and IV metastatic melanoma using 159 multiplexed mass cytometry-based imaging of protein markers and RNA transcripts, and found that CXCL9 and CXCL10 were spatially co-localized with checkpoint molecule-positive CD8+ T cells (17), and that upregulation of checkpoint molecules in these T cells made CXCL9/10 has been shown to serve as a new target for tumors in immunotherapy (18). In the bladder cancer samples of this study, we also found that CXCL9/10 expression positively correlated with both PD-L1 and TMB, which plays a key role for immunosuppressive therapy to work. Our analysis also showed that the AUC of CXCL9/10 mRNA was higher than that of PD-L1 for predicting response to treatment with Atezolizumab, suggesting that CXCL9/10 has a stronger ability to predict treatment response. Furthermore, high CXCL9/10 mRNA expression was found to be a predictor of better treatment outcome in the IMvigor210 dataset, resulting in longer survival in patients who received Atezolizumab treatment. The main objective of this study is to select molecular markers that can effectively predict the response to treatment with Atezolizumab in patients with bladder urothelial carcinoma, and the future development will be the combination of multiple molecular markers to benefit patients. The results of this study were analyzed using a bioinformatic approach, which were limited to the significance of mRNA expression, with confirmation at the protein level awaiting further validation. Immunotherapy has shown promising results in the treatment of muscle invasive bladder cancer (MIBC), both in neoadjuvant and adjuvant settings. However, there is still a lack of effective biomarkers to predict treatment efficacy (19,20). The potential of CXCL9/10 as a predictive biomarker for neoadjuvant and adjuvant immunotherapy in MIBC is promising and requires further validation.
Conclusions
We propose a novel genetic signature, which also seems to be able to reliably stratify the risk profile of bladder cancer patients, with prognostic significance. Moreover, our signature appears to be able to indicate which patients will show a better response to immunotherapy. These results could significantly improve the possibility of designing patient-oriented treatment strategies in the next future. Based on real-world datasets, CXCL9 and CXCL10 mRNA were found to be upregulated in bladder cancer patients who showed treatment response to Atezolizumab. Moreover, CXCL9 and CXCL10 mRNA expression was positively correlated with both PD-L1 and TMB. CXCL9 and CXCL10 mRNA seem to have a stronger ability than PD-L1 mRNA to predict responsiveness to Atezolizumab treatment. Patients with high CXCL9 and CXCL10 mRNA also show better OS in the Atezolizumab treated cohort. CXCL9 and CXCL10 mRNA can be used as molecular markers not only to predict treatment response, but also to acquire a prognostic value in bladder cancer patients treated with Atezolizumab.
Acknowledgments
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article:
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-220/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-220/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-220/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Flaig TW, Spiess PE, Abern M, et al. NCCN Guidelines® Insights: Bladder Cancer, Version 2.2022. J Natl Compr Canc Netw 2022;20:866-78. [Crossref] [PubMed]
- Mancini M, Righetto M, Noessner E. Checkpoint Inhibition in Bladder Cancer: Clinical Expectations, Current Evidence, and Proposal of Future Strategies Based on a Tumor-Specific Immunobiological Approach. Cancers (Basel) 2021;13:6016. [Crossref] [PubMed]
- Fitzsimmons TS, Singh N, Walker TDJ, et al. Immune checkpoint inhibitors efficacy across solid cancers and the utility of PD-L1 as a biomarker of response: a systematic review and meta-analysis. Front Med (Lausanne) 2023;10:1192762. [Crossref] [PubMed]
- Dall'Olio FG, Marabelle A, Caramella C, et al. Tumour burden and efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol 2022;19:75-90. [Crossref] [PubMed]
- Wu P, Cao Z, Wu S. New Progress of Epigenetic Biomarkers in Urological Cancer. Dis Markers 2016;2016:9864047. [Crossref] [PubMed]
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. [Crossref] [PubMed]
- Rose TL, Weir WH, Mayhew GM, et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br J Cancer 2021;125:1251-60. [Crossref] [PubMed]
- Paucek RD, Baltimore D, Li G. The Cellular Immunotherapy Revolution: Arming the Immune System for Precision Therapy. Trends Immunol 2019;40:292-309. [Crossref] [PubMed]
- Rizzo A, Ricci AD, Brandi G. PD-L1, TMB, MSI, and Other Predictors of Response to Immune Checkpoint Inhibitors in Biliary Tract Cancer. Cancers (Basel) 2021;13:558. [Crossref] [PubMed]
- Tong G, Zhu M, Chen Y, et al. Intratumoral CD8(+) T cells as a potential positive predictor of chemoimmunotherapy response in PD-L1-negative advanced gastric cancer patients: a retrospective cohort study. J Gastrointest Oncol 2022;13:1668-78. [Crossref] [PubMed]
- Aggen DH, Drake CG. Biomarkers for immunotherapy in bladder cancer: a moving target. J Immunother Cancer 2017;5:94. [Crossref] [PubMed]
- Tang Q, Chen Y, Li X, et al. The role of PD-1/PD-L1 and application of immune-checkpoint inhibitors in human cancers. Front Immunol 2022;13:964442. [Crossref] [PubMed]
- Kang K, Xie F, Mao J, et al. Significance of Tumor Mutation Burden in Immune Infiltration and Prognosis in Cutaneous Melanoma. Front Oncol 2020;10:573141. [Crossref] [PubMed]
- Ricciuti B, Wang X, Alessi JV, et al. Association of High Tumor Mutation Burden in Non-Small Cell Lung Cancers With Increased Immune Infiltration and Improved Clinical Outcomes of PD-L1 Blockade Across PD-L1 Expression Levels. JAMA Oncol 2022;8:1160-8. [Crossref] [PubMed]
- McGrail DJ, Pilié PG, Rashid NU, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 2021;32:661-72. [Crossref] [PubMed]
- Reschke R, Gajewski TF. CXCL9 and CXCL10 bring the heat to tumors. Sci Immunol 2022;7:eabq6509. [Crossref] [PubMed]
- Hoch T, Schulz D, Eling N, et al. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci Immunol 2022;7:eabk1692.
- House IG, Savas P, Lai J, et al. Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin Cancer Res 2020;26:487-504. [Crossref] [PubMed]
- Iacovino ML, Miceli CC, De Felice M, et al. Novel Therapeutic Opportunities in Neoadjuvant Setting in Urothelial Cancers: A New Horizon Opened by Molecular Classification and Immune Checkpoint Inhibitors. Int J Mol Sci 2022;23:1133. [Crossref] [PubMed]
- Barone B, Calogero A, Scafuri L, et al. Immune Checkpoint Inhibitors as a Neoadjuvant/Adjuvant Treatment of Muscle-Invasive Bladder Cancer: A Systematic Review. Cancers (Basel) 2022;14:2545. [Crossref] [PubMed]


