
Abnormal genetic and epigenetic patterns of m6A regulators associated with tumor microenvironment in colorectal cancer
Highlight box
Key findings
• Aberrant expression of m6A regulators in colorectal cancer is associated with mutation, CNV, and chromatin accessibility, owing to both genetic and epigenetic modifications.
What is known and what is new?
• The m6A methylation regulators played a vital role in affecting the prognosis of colorectal cancer.
• The genetic and epigenetic patterns, as well as tumor microenvironment infiltration characteristics of m6A regulators in colorectal cancer were revealed.
What is the implication, and what should change now?
• The molecular characteristics of m6A regulators may guide the development of more effective immunotherapy strategies in colorectal cancer.
Introduction
Colorectal cancer (CRC/COAD) is among the most frequently occurring human cancers worldwide, ranking third in incidence, and is the second leading cause of cancer-related deaths (1,2). Despite progress in surgery, chemoradiation, and targeted therapies, the survival rate of patients with CRC has not dramatically improved, owing to delayed diagnosis, rapid tumor progression, and ease of metastasis (3,4). Therefore, an extensive and in-depth understanding of the molecular characteristics of CRC is critical to enable early diagnosis and the development of therapeutic strategies and to improve patient prognosis.
As the most prominent mRNA modification, N6-methyladenosine (m6A) is widely found in mRNA, as well as in long non-coding RNA (lncRNA) and microRNA (miRNA) (5). In mammalian cells, m6A modification is a dynamic reversible process that is regulated by methyltransferases, demethylases, and binding proteins, also known as “writers”, “erasers”, and “readers” (6). The methyltransferases or writers include METTL3, METTL14, RBM15, RBM15B, KIAA1429 [also known as Vir like m6A methyltransferase associated (VIRMA)], WT1 associated protein (WTAP), CBLL1, and ZC3H13; the demethylases or erasers include FTO alpha-ketoglutarate dependent dioxygenase (FTO) and ALKBH5; and the main readers are YTHDF1/2/3, YTHDC1/2, HNRNPA2B1, Leucine rich pentatricopeptide repeat containing (LRPPRC), and FMR1 (7-9). The m6A regulators have crucial roles in many fundamental biological processes, including RNA splicing, translation efficiency, and mRNA stability (10). Dysregulation of expression and genetic changes of m6A regulators are related to various disease processes, including adipogenesis, impaired cell differentiation, malignant tumor progression, and immunomodulatory abnormalities (11).
Although immunotherapy using immunological checkpoint blockade [programmed cell death 1/programmed death ligand 1 (PD-1/L1) and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4)] has shown considerable clinical efficacy in a small percentage of patients, the majority of patients experienced minimal clinical benefit; thus, new techniques are urgently required to meet clinical needs. Tumor progression is a multistep process. Traditionally, research in this field has focused on the genetic and epigenetic variation in tumor cells. However, the tumor microenvironment (TME) also has a critical role in tumor progression and is conducive to tumor cell growth and survival. Besides tumor cells, the complex TME contains stromal cells (cancer-associated fibroblasts, macrophages, etc.), distant recruited cells (myeloid cells, lymphocytes, etc.), secreted factors (chemokines, cytokines, growth factors, etc.), and new blood vessels (12). Tumor cells interact directly and indirectly with TME components and change their biological behavior (for instance, by inducing proliferation, angiogenesis, and immune tolerance), indicating a critical role of the TME in tumor progression and immune escape (13,14). Notably, the characteristics of TME cell infiltration have been shown to predict response to immunotherapy, thereby increasing the success of novel immunotherapeutic strategies (15). Therefore, determining the comprehensive landscape of TME characteristics and finding promising biomarkers will be an effective means of identifying new therapeutic targets. Although accumulating evidence indicates that aberrant m6A modifications are necessary for tumor progression in various cancers, only a few studies have considered their role in CRC (16,17). Thus, the molecular patterns and TME infiltration characteristics of m6A regulators in CRC remain largely unknown.
Here, we revealed that m6A regulators were frequently dysregulated in CRC and associated with mutations, copy number variations (CNVs), DNA methylation, and changes in chromatin accessibility. The expression of most m6A regulators was positively correlated with TME infiltration, including that of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. The molecular patterns and TME infiltration characteristics of m6A regulators in CRC described here might be critical for developing immunotherapeutic strategies and improving prognosis. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-186/rc).
Methods
Datasets
Level 3 raw count data consisting of mutations, CNV, and relevant clinical data for 534 CRC patients were obtained from cBioPortal for Cancer Genomics (18). Of these, 380 patients had RNA-seq data and 441 patients had DNA methylation. The mRNA expression profiles and DNA methylation data of these tumor tissues and 51 control samples were obtained from The Cancer Genome Atlas (TCGA) via the University of California Santa Cruz (UCSC) Xena (19). The assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq) data of COAD was obtained from https://gdc.cancer.gov/about-data/publications/ATACseq-AWG (20).
Expression, mutation, and CNV analysis
The mRNA expression profiles of CRC tissues and control samples were obtained from TCGA via UCSC Xena (19). The data analysis is based on the UCSC Xena website. Briefly, the gene expression profile was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform by the University of North Carolina TCGA genome characterization center. Gene expression was estimated as in log2 (x+1) transformed RNAseq by Expectation Maximization (RSEM) normalized count.
The mutation analysis was performed using cBioPortal for Cancer Genomics based on data from studies of TCGA, Pan-Cancer Atlas (18,21). Analysis of CNV followed the TCGA publication guidelines (http://cancergenome.nih.gov/publications) (22). Each sample’s copy number was normalized, and estimates of the mean copy number for segments overlapping the whole genome were obtained for analysis (22). Genes were categorized using the mean cut-offs of the GISTIC (genomic identification of significant targets in cancer) algorithm. The frequency of amplification or deletion was calculated by the following formula: (number of samples with amplification or deletion in a group)/(total number of samples in a group). Copy number calls were categorized as amplifications or deletions using thresholds of ≥1 for amplification and ≤−1 for deletion.
Methylation and ATAC-seq analysis
DNA methylation profiles of 441 CRC patients were obtained using the Infinium HumanMethylation450 array from the Genome Data Commons (GDC) Data Portal. Beta values were derived at the Johns Hopkins University and University of Southern California TCGA genome characterization center. DNA methylation values, described as beta values, are recorded for each array probe in each sample via BeadStudio software (23).
The chromatin accessibility of each gene was calculated based on all peaks, including promoter peaks and enhancer peaks. As for ATAC-seq analysis, normalized count matrix: a prior count of 5 is added to the raw counts, then put into a “counts per million”, then log2 transformed, then quantile normalized. The values were averaged [i.e., log2((count+5)PM)-qn values] across all technical replicates and all biospecimens belong to the same TCGA sample (20). The relationships among mRNA expression, somatic mutations, CNV, DNA methylation, and ATAC-seq were analyzed using the UCSC Xena browser (https://xenabrowser.net/heatmap/) and cBioPortal for Cancer Genomics.
Microenvironment infiltration analysis
The microenvironment infiltration analysis was performed using Tumor Immune Estimation Resource (TIMER), which is a comprehensive resource for systematic analysis of immune infiltrates across diverse cancer types (24). We analyzed the correlations of expression of m6A regulators with the abundance of immune infiltrates in COAD, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. Besides, the abundance of tumor-infiltrating immune cells was explored according to somatic mutations (ZC3H13) and CNV (YTHDC2, YTHDF1, YTHDF3, VIRMA, and ZC3H13). Expression scatter plots with Spearman’s correlation and estimated statistical significance were generated for m6A regulators. The gene expression level was displayed as log2 RSEM.
Statistical analysis
The differences in gene expression between the two groups were evaluated using a two-tailed Mann-Whitney U-test. Correlation analysis was performed using the Pearson coefficient. The Kaplan-Meier method was used for the bilateral logarithmic rank test in the overall survival analysis. Statistical analyses were performed using GraphPad Prism software. For all tests, P values were two-sided; P<0.05 with Bonferroni correction was considered to indicate statistical significance.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Expression of m6A regulators was frequently dysregulated in CRC
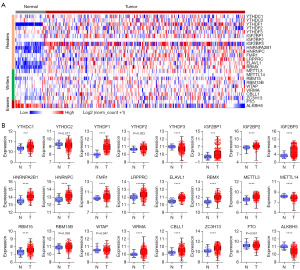
To determine the molecular characteristics of m6A regulators in CRC, we first analyzed the expression patterns of m6A regulators in TCGA CRC samples (51 normal vs. 380 primary tumors) via UCSC Xena. Interestingly, the expression of m6A regulators was frequently dysregulated in CRC (Figure 1A). Among them, 16 m6A regulators were significantly upregulated in CRC compared with in para-carcinoma tissues, including YTHDC1, YTHDF1, IGF2BP1/2/3, and HNRNPA2B1, etc. (Figure 1B). However, only YTHDF3, METTL14, and ALKBH5 were significantly downregulated in CRC (Figure 1B). Moreover, the expression of 5 m6A regulators (YTHDC2, YTHDF2, RBM15B, WTAP, and FTO) showed no difference between normal and tumor samples (Figure 1B). These results indicated that m6A regulators were frequently dysregulated in CRC.
The mutation characteristic of m6A regulators in CRC
To determine whether genetic alterations affect the expressions of m6A regulators in CRC, we first used cBioPortal to identify somatic mutations in 534 CRC tumor samples. All of the m6A regulators had somatic mutations, with frequencies ranging from 0.7% to 7% (Figure 2A). Only three had a mutation frequency of less than 1%, namely HNRNPA2B1 (0.9%), HNRNPC (0.9%), and ALKBH5 (0.7%); and five had a mutation frequency greater than 3%, namely YTHDC2 (5.0%), IGF2BP1 (4.0%), RBM15 (4.0%), VIRMA (5.0%), and ZC3H13 (7.0%) (Figure 2A). Notably, ZC3H13 was the most frequently mutated among these m6A regulators in CRC, with a mutation frequency of 7.0% (Figure 2A).
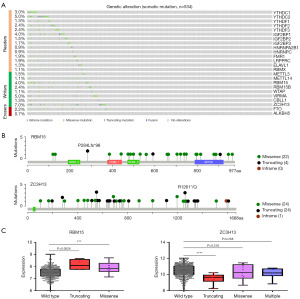
To determine the associations between the mutations and the expression of m6A regulators, we compared the expression levels of m6A regulators between the mutation group and the wild-type group. Interestingly, the expression of ZC3H13 was significantly downregulated in the mutation group compared with the wild-type group (Figure S1). However, RBM15 expression was upregulated in the mutation group (Figure S1), suggesting that the mutations in RBM15 might activate their expression.
Since different types of mutations have different effects on gene expression, we analyzed the types of mutations of RBM15 and ZC3H13 that occurred in CRC. Interestingly, both RBM15 and ZC3H13 had missense mutations, along with varying degrees of truncating (Figure 2B). Moreover, the expression of RBM15 and ZC3H13 in the truncating group was different from that of the wild type (Figure 2C). These data reveal that somatic mutation account only partially for the expression patterns of m6A regulators in CRC.
CNV of m6A regulators inordinately co-occur in CRC and are positively correlated with expression
Next, we analyzed CNV in 534 TCGA CRC tumor samples via cBioPortal. Compared with somatic cell mutations, CNV occurred at a relatively low frequency in CRC, with only five genes having a CNV frequency greater than 1%, namely YTHDC2 (2.4%), YTHDF1 (7.0%), YTHDF3 (1.9%), VIRMA (1.7%), and ZC3H13 (3.0%) (Figure 3A).
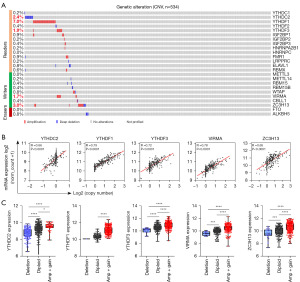
To determine the associations between CNV and the expression of m6A regulators, a correlation analysis was performed based on the TCGA CRC samples with both gene expression and copy number information. The correlation analysis focused on YTHDC2, YTHDF1, YTHDF3, VIRMA, and ZC3H13 owing to their CNV frequencies being greater than 1% (Figure 3A). As anticipated, a significant positive correlation was found between mRNA expression and copy number in CRC (Figure 3B). We further investigated the mRNA expression in the deletion (deep deletion and shallow deletion), diploid, and Amp + Gain (amplification and gain) groups. Compared with the diploid group, the expression of these five genes (YTHDC2, YTHDF1, YTHDF3, VIRMA, and ZC3H13) was significantly downregulated in the deletion group, whereas it was significantly upregulated in the Amp + Gain group (Figure 3C). These results suggest that copy number has a great influence on gene expression.
DNA methylation did not associate with m6A regulators’ expression
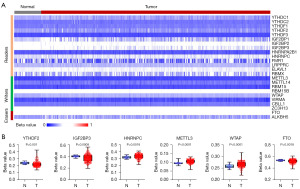
As the main form of epigenetic modification, DNA methylation has an important role in tumor progression (25). We next analyzed the DNA methylation patterns of m6A regulators in 441 TCGA CRC samples via UCSC Xena. Notably, YTHDF2, IGF2BP3, and FTO were hypomethylated in CRC, whereas HNRNPC, METTL3, and WTAP remained hypermethylation in CRC (Figure 4A,4B and Figure S2). Unfortunately, there was no correlation between the methylation level of 24 m6A regulators and their gene expression (data not shown).
Chromatin accessibility associated with m6A regulators’ expression
Upon further investigation of the molecular characteristics of m6A regulators in CRC, we unexpectedly noted that the promoter and enhancer regions of most m6A regulators (relative to YTHDC1, YTHDC2, FMR1, FTO, etc.) maintained high chromatin accessibility along with high mRNA expression (Figure 5A). Indeed, the chromatin accessibility of YTHDC1, YTHDC2, YTHDF3, FMR1, RBMX, ZC3H13, and FTO tended to be the low state with low mRNA expression (Figure 5A). These results suggest that chromatin accessibility might influence the gene expression of these regulators.
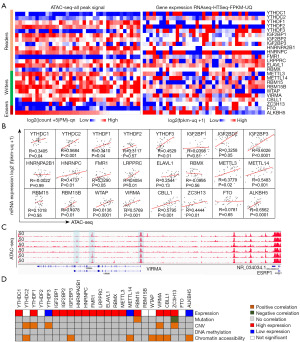
To evaluate whether any of these open chromatin regions had a regulatory effect on gene expression, we re-analyzed the correlations between regulatory elements and target genes with respect to chromatin accessibility using TCGA COAD ATAC-seq with matched RNA sequencing (20). Interestingly, positive correlations between these chromatin regions and target gene expression were found for 7 m6A regulators, including YTHDC2, WTAP, ALKBH5, etc. (Figure 5B). However, there were also some m6A regulators for which there was no correlation between chromatin regions and target gene expression; these included YTHDF2, IGF2BP1, HNRNPA2B1, ELAVL1, RBMX, RBM15, and FTO, etc. (Figure 5B). VIRMA was a case in which the promoter and enhancer regions maintained high chromatin accessibility (Figure 5C).
Next, we comprehensively analyzed the effects of mutations, CNV, methylation, and chromatin accessibility on gene expression. Of the 19 dysregulated m6A regulators, 7 had positive correlations between chromatin accessibility and target gene expression (Figure 5D), suggesting that chromatin accessibility might have a major effect on gene expression. Moreover, mutations of RBM15 and ZC3H13 and CNV of YTHDC2, YTHDF1, YTHDF3, VIRMA, and C3H13 affected gene expression (Figure 5D). These results suggest that gene expression of dysregulated m6A regulators is influenced by genetics (mutations and CNV) and epigenetics (chromatin accessibility).
Expression of m6A regulators is positively correlated with TME infiltration in COAD
As tumor-infiltrating lymphocytes are independent predictors of lymph node status and survival in cancers (26,27), we investigated whether the expression of m6A regulators was correlated with immune infiltration levels in COAD via TIMER. Notably, the expression levels of YTHDC2, IGF2BP3, WTAP, and ALKBH5 had negative correlations with tumor purity in COAD, whereas those of LRPPRC and RBMX had positive correlations with tumor purity (Figure 6A). Moreover, the expression levels of most m6A regulators had significant positive correlations with levels of infiltrating B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Figure 6A). Of these m6A regulators, 18 genes were positively associated with B cell infiltration levels, 20 genes were associated with CD8+ T cell, neutrophil, and CD4+ T cell infiltration levels, 19 genes were associated with macrophage infiltration levels, and 22 genes were associated with dendritic cell infiltration levels (Figure 6A). These findings strongly suggest that m6A regulators have a pivotal role in immune infiltration in COAD.
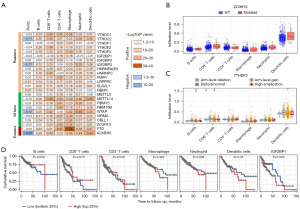
As ZC3H13 was the most frequently mutated m6A regulator in COAD, we next analyzed the association of ZC3H13 mutations with immune infiltration levels in the diverse types group. The infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells were not significantly different between the mutant and wild-type groups (Figure 6B). However, it is worth noting that the infiltration levels of B cells were significantly different among the diverse type groups according to the YTHDF3 copy number (Figure 6C). Consistently, similar results were found for the other four m6A regulators (YTHDC2, YTHDF1, VIRMA, and ZC3H13) (Figure S3).
Next, we investigated the potential prognostic value of the infiltration levels of different immune cell types in COAD via TIMER. The infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells were not significantly correlated with OS in COAD (Figure 6D). However, the expression of IGF2BP1 was significantly correlated with OS (Figure 6D), suggesting that it might be a good prognostic marker for COAD patients.
Discussion
Accumulating evidence demonstrates that m6A mRNA modification represents a new layer of post-transcriptional gene regulation that is widely found in mRNAs, lncRNAs, and miRNAs (28-30). These m6A modifications participate in almost every aspect of RNA metabolism, including RNA splicing, subcellular localization, translation, mRNA stability, and RNA-protein interactions (31-33). Accompanied by the dynamic and delicate interplay between m6A methyltransferases and demethylases, m6A modifications play crucial parts in cell fate determination, sex determination, pluripotency, and cancer progression (34,35). However, the molecular characterization and clinical significance of m6A modifications in human cancers, especially CRC, are just beginning to be explored. In CRC, other researchers provided a comprehensive analysis of four RNA modifications, identified m6A-related mRNA biomarkers related to the clinicopathology and prognosis, and even developed an m6Ascore to predict performance for overall survival and clinical efficacy of immunotherapy (36-38). These studies mainly focused on the influence of m6A regulators themselves on CRC. In our study, we pay more attention to the upstream regulation mechanism of the expression of m6A regulators. Thus, our study provides insight into the potential role of m6A regulators in tumor immunology, which will have benefits in terms of prognosis and immunotherapy.
Genetic changes caused by somatic mutation and CNV have an important role in cancer and are probably potential biomarkers for diagnosis and prognosis (39-41). Recently, somatic mutations and CNV were found to activate or silence many novel genes that contribute to human cancer (42). Here, we found that all the m6A regulators had somatic mutations in CRC (Figure 2A). Mutations of RBM15 were more likely to activate their expression, whereas mutations of ZC3H13 were more likely to inhibit its expression. However, some m6A regulators had a high frequency of mutations with no corresponding effect on their expression. Besides genetic mutations, CNV often occur in oncogenic signaling pathways in tumors, including in MYC, Hippo, Phosphatidylinositol 3-kinase/Akt kinase (PI3K/AKT), Receptor tyrosine kinases-rat sarcoma virus (RTK-RAS), Transforming Growth Factor beta (TGF-β) signaling, and Wnt/β-catenin signaling (41). Compared with somatic mutations, CNV of m6A regulators occur at a relatively low frequency in CRC. However, a significant positive correlation was found between mRNA expression and copy number in these m6A regulators (Figure 3). Despite the low frequency of CNV in m6A regulators, it was sufficient to affect their gene expression, suggesting that CNV play an important part in the activation of m6A regulators.
As a complement to genetic changes, epigenetic factors including DNA methylation have an integral role in tumorigenesis (25,43-45). Epigenetic silencing by DNA methylation has an important effect on many critical genes (25,43-45). Here, we found that three m6A regulators (YTHDF2, IGF2BP3, and FTO) were hypomethylated in CRC, whereas HNRNPC, METTL3 and WTAP remained hypermethylation in CRC (Figure 4). This suggests that DNA methylation may be involved in gene expression. However, there was no correlation between the methylation level of 24 m6A regulators and their gene expression, suggesting DNA methylation might not be the factor causing differences in the gene expression of m6A regulators.
Abnormal gene expression patterns allow cancer cells to acquire their hallmark characteristics, while genomic instability along with genetic alterations (such as somatic mutations and CNV) enables tumorigenesis (39-41). Chromatin accessibility, a key factor in transcription regulation, has a central role in gene expression. Therefore, we analyzed the chromatin accessibility of m6A regulators based on ATAC-seq from TCGA. Interestingly, the promoter and enhancer regions of most m6A regulators maintained high chromatin accessibility with high mRNA expression (Figure 5). Moreover, positive correlations between these chromatin regions and the expression of target genes were found across 7 m6A regulators (Figure 5). These results suggest that chromatin accessibility affected the expression of m6A regulators. However, the contribution of chromatin accessibility is highly speculative in this present state, which needs to be further explored.
To date, the effects of m6A regulators on immune infiltration in CRC have not been fully elucidated. Several studies have shown that m6A regulators participate in immune infiltration in gastric cancer, head and neck squamous cell carcinoma, nasopharyngeal carcinoma, and lung cancer (46-50), suggesting that they influence tumor immunity to a certain extent. Here, we demonstrated significant positive correlations between the infiltration levels of CD8+ and CD4+ T cells, macrophages, dendritic cells, and neutrophils and the expression levels of most m6A regulators in COAD. These findings strongly suggest that m6A regulators have a pivotal role in the recruitment and regulation of immune infiltrating cells in COAD. Moreover, the infiltration levels of B cells, CD8+ T cells, and dendritic cells showed significant differences among the diverse type groups according to YTHDF3, YTHDC2, YTHDF1, VIRMA, and ZC3H13 copy number, but not for diverse type groups according to ZC3H13 mutation. This suggests that the ZC3H13 mutation had no significant impact on tumor immunity. It is possible that dysregulated m6A regulators in the infiltrating immune cells drive the differences in the expression of m6A genes in the subset of tumors that exhibit high levels of immune cell infiltration, which need to be further explored. Moreover, infiltration levels of immune cells (CD8+ and CD4+ T cells, macrophages, dendritic cells, and neutrophils) had no significant association with patient survival, mainly because patients’ immune cell infiltration levels did not change much during treatment. Despite this, the expression of IGF2BP1 was significantly correlated with OS (Figure 6D), suggesting that IGF2BP1 might be a good prognostic marker in COAD patients.
Conclusions
In summary, m6A regulators were frequently dysregulated in CRC partly due to genetics (CNV) and epigenetics (chromatin accessibility). The increased expression of most m6A regulators was correlated with increased immune infiltration levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in COAD. Therefore, m6A regulators are likely to play an important part in immune cell infiltration and have the potential as novel prognostic molecular markers and therapeutic targets.
Acknowledgments
We thank the authors of TCGA, cBioPortal, and GEO for providing available data.
Funding: This work was funded by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-186/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-186/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-186/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond) 2021;41:1037-48. [Crossref] [PubMed]
- Ciardiello F, Ciardiello D, Martini G, et al. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin 2022;72:372-401. [Crossref] [PubMed]
- Cervantes A, Adam R, Roselló S, et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol 2023;34:10-32. [Crossref] [PubMed]
- Li J, Zhang H, Wang H N. (1)-methyladenosine modification in cancer biology: Current status and future perspectives. Comput Struct Biotechnol J 2022;20:6578-85. [Crossref] [PubMed]
- Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. [Crossref] [PubMed]
- Tong J, Flavell RA, Li HB. RNA m6A modification and its function in diseases. Front Med 2018;12:481-9. [Crossref] [PubMed]
- Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. [Crossref] [PubMed]
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017;169:824-35.e14. [Crossref] [PubMed]
- Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell 2023;83:428-41. [Crossref] [PubMed]
- Cui L, Ma R, Cai J, et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct Target Ther 2022;7:334. [Crossref] [PubMed]
- Demaria O, Cornen S, Daëron M, et al. Publisher Correction: Harnessing innate immunity in cancer therapy. Nature 2019;576:E3. [Crossref] [PubMed]
- de Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023;41:374-403. [Crossref] [PubMed]
- Tiwari A, Trivedi R, Lin SY. Tumor microenvironment: barrier or opportunity towards effective cancer therapy. J Biomed Sci 2022;29:83. [Crossref] [PubMed]
- Kirchhammer N, Trefny MP, Auf der Maur P, et al. Combination cancer immunotherapies: Emerging treatment strategies adapted to the tumor microenvironment. Sci Transl Med 2022;14:eabo3605. [Crossref] [PubMed]
- Liu T, Li C, Jin L, et al. The Prognostic Value of m6A RNA Methylation Regulators in Colon Adenocarcinoma. Med Sci Monit 2019;25:9435-45. [Crossref] [PubMed]
- Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer 2019;18:112. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Corces MR, Granja JM, Shams S, et al. The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898.
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 2017;45:e22. [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun 2007;7:4. [PubMed]
- Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 2012;30:2678-83. [Crossref] [PubMed]
- Alarcón CR, Lee H, Goodarzi H, et al. N6-methyladenosine marks primary microRNAs for processing. Nature 2015;519:482-5. [Crossref] [PubMed]
- Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016;537:369-73. [Crossref] [PubMed]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31-42. [Crossref] [PubMed]
- Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science 2016;352:1408-12. [Crossref] [PubMed]
- Roignant JY, Soller M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet 2017;33:380-90. [Crossref] [PubMed]
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 2014;15:313-26. [Crossref] [PubMed]
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613. [Crossref] [PubMed]
- Pan Y, Ma P, Liu Y, et al. Multiple functions of m(6)A RNA methylation in cancer. J Hematol Oncol 2018;11:48. [Crossref] [PubMed]
- Chen H, Yao J, Bao R, et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer 2021;20:29. [Crossref] [PubMed]
- Zhang Z, Zhang X. Identification of m6A-Related Biomarkers Associated with Prognosis of Colorectal Cancer. Med Sci Monit 2021;27:e932370. [Crossref] [PubMed]
- Yue Q, Zhang Y, Wang F, et al. Characterization of m6A Methylation Modification Patterns in Colorectal Cancer Determines Prognosis and Tumor Microenvironment Infiltration. J Immunol Res 2022;2022:8766735. [Crossref] [PubMed]
- Li Y, Kang K, Krahn JM, et al. A comprehensive genomic pan-cancer classification using The Cancer Genome Atlas gene expression data. BMC Genomics 2017;18:508. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-16.e11. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-37.e10. [Crossref] [PubMed]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;173:371-85.e18. [Crossref] [PubMed]
- A Convergence of Genetics and Epigenetics in Cancer. Cell 2017;168:561-3. [Crossref] [PubMed]
- Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A 2017;114:7414-9. [Crossref] [PubMed]
- Saghafinia S, Mina M, Riggi N, et al. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep 2018;25:1066-80.e8. [Crossref] [PubMed]
- Zhang B, Wu Q, Li B, et al. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer 2020;19:53. [Crossref] [PubMed]
- Yi L, Wu G, Guo L, et al. Comprehensive Analysis of the PD-L1 and Immune Infiltrates of m(6)A RNA Methylation Regulators in Head and Neck Squamous Cell Carcinoma. Mol Ther Nucleic Acids 2020;21:299-314. [Crossref] [PubMed]
- Xu F, Zhang H, Chen J, et al. Immune signature of T follicular helper cells predicts clinical prognostic and therapeutic impact in lung squamous cell carcinoma. Int Immunopharmacol 2020;81:105932. [Crossref] [PubMed]
- Xu F, Chen JX, Yang XB, et al. Analysis of Lung Adenocarcinoma Subtypes Based on Immune Signatures Identifies Clinical Implications for Cancer Therapy. Mol Ther Oncolytics 2020;17:241-9. [Crossref] [PubMed]
- Lu S, Yu Z, Xiao Z, et al. Gene Signatures and Prognostic Values of m(6)A Genes in Nasopharyngeal Carcinoma. Front Oncol 2020;10:875. [Crossref] [PubMed]


