
Combining Bulk RNA-seq and scRNA-seq data to identify RNA m5C methyltransferases NSUN1: a rising star as a biomarker for cancer diagnosis, prognosis and therapy
Highlight box
Key findings
• NSUN1 may be an effective biomarker for early cancer diagnosis, prognosis and therapy.
What is known and what is new?
• The expression of NSUN1 is elevated in breast and colon cancer and can predict poor prognosis.
• Elevated NSUN1 expression have been observed in most human cancers. High NSUN1 expression indicates poor overall survival and disease-free survival.
What is the implication, and what should change now?
• These results herein imply that NSUN1 may be an effective biomarker for early cancer diagnosis, prognosis and therapy.
IntroductionOther Section
RNA 5-methylcytosine (m5C) methyltransferases NSUN1, also known as NOP2 and P120, is a member of the NOP2/SUN (NSUN) RNA methyltransferase family, which includes seven members: NSUN1-7 (1-4). NSUN1 is a nucleolar RNA-binding protein located on the chromosome 12p13.31 (5). Studies have found that NSUN1 is involved in the positive regulation of cell population proliferation, the regulation of signal transduction by p53 class mediators, and the assembly of large ribosomal subunits (6,7).
It is noteworthy that NSUN1 has been reported in six human cancers. The expression of NSUN1 was significantly higher in cancer tissues than in the corresponding tumor adjacent normal tissues, as in the case of breast cancer (BRCA) (8), colon adenocarcinoma (COAD) (9), stomach adenocarcinoma (STAD) (10), prostate adenocarcinoma (PRAD) (11,12), rectum adenocarcinoma (READ) (13), and hepatocellular cancer (HCC) (14). It can promote the proliferation, invasion, migration of cancer cells in vitro and promote tumor growth and metastasis in vivo (10,14). In addition, it was found that long noncoding RNA (lncRNA)-hPVT1 promoted HCC cell proliferation, cell cycle and acquired stem cell-like properties by stabilizing NSUN1 protein. The regulation of lncRNA-hPVT1/NSUN1 pathway may have a positive effect on the treatment of HCC (14). The study also found that NSUN1 gene expression was significantly associated with microsatellite instability (MSI), tumor mutation burden (TMB) and immunity in renal cancer, and elevated expression was associated with poor overall survival (OS) (13). These results suggest that NSUN1 may be a potential prognostic factor for OS and a molecular marker for immunotherapy.
To gain a more complete and comprehensive understanding of the NSUN1 molecule and its role in various cancers, the expression, localization, variation, epigenetic modification and biological function of NSUN1 in human cancers were analyzed herein. This study also characterized the expression of NSUN1 in different immune cell types in the tumor immune microenvironment using single-cell RNA sequencing (scRNA-seq) data from public databases. The analysis herein provided evidence that NSUN1 could be used as a potential diagnostic, prognostic, and therapeutic biomarker for cancer. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-66/rc).
MethodsOther Section
Gene expression analysis using Bulk RNA-seq and scRNA-seq data
Gene expression in different tumor and normal tissues was analyzed using Tumor Immune Estimation Resource (TIMER2; http://timer.cistrome.org/). The Cancer Genome Atlas (TCGA) provided the data source of the public dataset. Receiver operating characteristic (ROC) curve (https://www.xiantao.love/) was drawn by using Xiantao tools. scRNA-seq and immunohistochemistry data were fetched from The Human Protein Atlas (HPA; https://www.proteinatlas.org/) website, and mapping was performed to demonstrate the expression of NSUN1 in different cell types.
TMB, MSI and immune checkpoints expression analysis
Studying the relationship between the expression of NSUN1 and immune checkpoints [programmed death-1 (PD1), programmed cell death-ligand 1 (PDL1), cytotoxic T-lymphocyte antigen 4 (CTLA4), etc.] using Assistant for Clinical Bioinformatics (ACLBI; https://www.aclbi.com). Using the ACLBI was applied to study the relationship between NSUN1 expression and TMB/MSI in various cancers. “Immune-related” and “Mutation Analysis” function were used in ACLBI tool. The parameter filled in “sample” was “all tumors”.
Gene location and alteration analysis
The HPA was used to identify the gene location. “SUBCELLULAR” and “HUMAN CELLS” function were used in HPA database. Immunofluorescence staining was used to observe the localization of NSUN1 protein in A-431, U-2 OS and U251 MG cells. Immunohistochemical staining images were also obtained from the HPA database. The cBioPortal tool (http://www.cbioportal.org/) was used to analyze the alteration frequency and mutation type (15,16). “TCGA PanCancer Atlas Studies” was selected to analysis by “Query By Gene”. The parameter filled in “Enter Genes” was “NOP2”.
Gene methylation and phosphorylation characteristics analysis
The University of ALabama at Birmingham CANcer data analysis Portal (UCLCAN; http://ualcan.path.uab.edu/index.html) was used to analyze the epigenetic profile of NSUN1. Genomic and proteomic data were obtained from the TCGA and Clinical Proteomic Tumor Analysis Consortium (CPTAC; https://cptac-data-portal.georgetown.edu/cptacPublic/).
Survival analysis
The relationship between NSUN1 expression and OS and disease-free survival (DFS) was analyzed using Gene Expression Profiling Interactive Analysis 2 (GEPIA2) (17). High and low expression groups were split using the median as the expression threshold. Survival plots use Cox proportional hazard ratio and 95% confidence interval information. Hypothesis testing was performed using the log-rank test.
Enrichment analysis
The top100 genes significantly correlated with NSUN1 expression were found by GEPIA2 analysis. The R package of “clusterProfiler”, “enrichplot” and “ggplot2” were used to perform the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment and visualization.
Statistical analysis
Data and figures for the article was obtained from public databases (Table S1). GO and KEGG pathway enrichment analyses were performed using R software (Version 4.0.2). This study was conducted in compliance with the Helsinki Declaration (as revised in 2013).
ResultsOther Section
NSUN1 is highly expressed in cancer
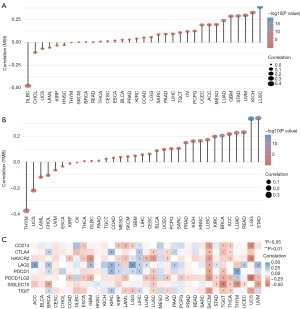
We analyzed the expression of NSUN1 in various tumors using Bulk RNA-seq data, and found that the expression of NSUN1 increased significantly in most tumors, and it was more significant in BRCA, cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC) (Figure 1A, Figure S1). The increased NSUN1 expression was also confirmed at protein level (Figure 1B, Figure S2A). Furthermore, the ROC curve was used to evaluate the sensitivity and specificity of NSUN1 in predicting cancer, and the results confirmed the important value of NSUN1 in predicting cancer (Figure S2B). This finding has important clinical value in the early diagnosis of cancer. NSUN1 may be a promising biomarker for the early and accurate diagnosis.

scRNA-seq is widely used in cancer research, and its ability to analyze specific molecular signatures of different cell groups is of great value in revealing tumor heterogeneity. Understanding the infiltration characteristics of immune cells in the tumor immune microenvironment is of great significance for exploring new immune checkpoints and guiding the application of immunotherapy drugs. Using scRNA-seq data from public databases, we found that NSUN1 expression was significantly high in immune cells than in other cell types in breast, liver, lung, stomach, lymph node, endometrium, and bone marrow (Figure 2A-2G). For example, in breast, the expression of NSUN1 was higher in T cells and dendritic cells (DCs) (Figure 2A). In liver, stomach and lymph nodes, NSUN1 expression was higher in B cells and T cells (Figure 2B,2F,2G). These findings suggest that NSUN1 may be more involved in immune regulation in these organs. In cancers derived from these organs, NSUN1 may play a more important role in immune regulation.

The expression of NSUN1 was highly correlated with the expression of TMB, MSI and immune checkpoints
TMB and MSI status were closely related to immunotherapy efficacy. Microsatellite-stable (MSS) tumors with high tumor mutational burden (TMB-H) benefit from immunotherapy. We analyzed TMB, MSI and immune checkpoints expression in cancers. NSUN1 expression was positively correlated with MSI in most cancers, except for diffuse large B-cell lymphoma (DLBC), CHOL, uterine carcinosarcoma (UCS), acute myeloid leukemia (LAML) and head and neck squamous cell carcinoma (HNSC) (Figure 3A). Moreover, we also observed a positive correlation between NSUN1 expression and TMB in most cancers, except for thymoma (THYM), UCS, LAML, CHOL, uveal melanoma (UVM), ESCA and kidney renal papillary cell carcinoma (KIRP) (Figure 3B). Interestingly, we found that expression of immune checkpoints-related genes was associated with NSUN1 expression, especially in skin cutaneous melanoma (SKCM), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), UCS, LIHC and kidney renal clear cell carcinoma (KIRC) (Figure 3C). It remains to be studied whether NSUN1 expression may affect the efficacy of immunotherapy. In addition, we researched the relationship between NSUN1 expression and immune cells in the tumor microenvironment. We found that NSUN1 expression was negatively associated with the number of infiltrating immune cells in some cancers, such as COAD, lung adenocarcinoma (LUAD), LUSC, SKCM, STAD and TGCT, suggesting that NSUN1 might play a role in immune evasion (Figure S3).
Location, variation, methylation, and phosphorylation characteristics of NSUN1
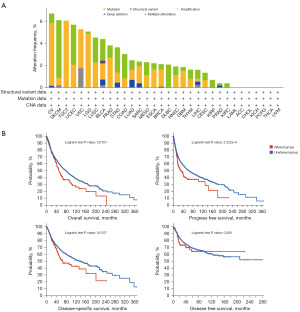
Immunofluorescence staining was used to detect NSUN1 subcellular distribution within the nucleus, endoplasmic reticulum (ER), and microtubules of A-431, U-2, U-251 cells. The result showed that the NSUN1 was located in the nucleus (Figure 4). The DNA alteration of NSUN1 was mainly amplification and mutation in TCGA pan-cancer. The frequency of NSUN1 alteration (>6%) was the highest in ovarian serous cystadenocarcinoma (OV) with “amplification” as the primary type (Figure 5A). In the log-rank test, patients in altered group did significantly worse in OS (P=0.0151), progression-free survival (PFS; P<0.0001) and disease-specific survival (DSS; P=0.0127) (Figure 5B). These data suggested that genetic variation of NSUN1 may be important prognostic factors for cancer patients.

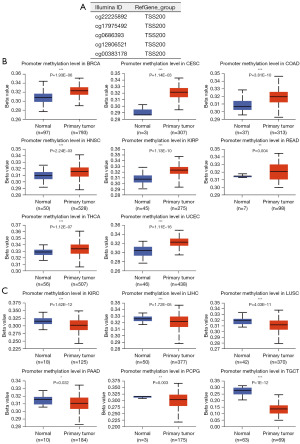
Phosphorylation is a critical signaling process for cell biological behavior. Interestingly, the present study found that phosphorylation of NSUN1 protein was significantly increased in tumor tissues compared with normal samples. Results from different cancers and different phosphorylation sites were reproducible and consistent (Figure 6, Figure S4). The results suggest that the phosphorylation modification of NSUN1 may be an important factor in the regulation of gene function. To clarify epigenetic characteristics of NSUN1, five probes in promoter were used for detecting DNA methylation level of NSUN1 (Figure 7A). This result suggests that the DNA methylation levels increased in BRCA, COAD, THCA etc., while decreased in KIRC, LIHC etc. (Figure 7B,7C). Our analysis shows that, across different cancers, promoter methylation levels are inconsistent with NSUN1 expression.

High expression of NSUN1 predicts poor prognosis
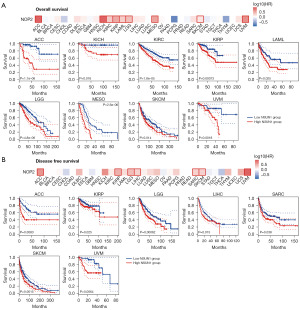
We analyzed the impact of NSUN1 on clinical prognosis. It was found that NSUN1 has good predictive value for OS and DFS in cancer patients. In adrenocortical carcinoma (ACC), kidney chromophobe (KICH), KIRC, KIRP, LAML, brain lower grade glioma (LGG), mesothelioma (MESO), SKCM and UVM, patients with high NSUN1 expression had worse OS. In ACC, KIRP, LGG, LIHC, sarcoma (SARC), SKCM and UVM, patients with high NSUN1 expression had worse DFS (Figure 8). These results suggest that NSUN1 may be an effective biomarker for prognosis.
GO and KEGG enrichment
Finally, we used the TCGA dataset to obtain 100 genes that were significantly correlated with NSUN1 expression through correlation analysis, and performed enrichment analysis. GO enrichment analysis showed that NSUN1-related genes were significantly related to ribosome biogenesis, DNA replication, rRNA processing and other biological processes (Figure 9A,9B). The locations of action were mainly chromosomes and ribosomes (Figure 9C,9D). NSUN1 can bind to DNA/RNA to exert biological functions such as helicase and catalytic activity (Figure 9E,9F). In addition, KEGG enrichment analysis found that NSUN1 may also be related to biological behaviors such as cell cycle and mismatch repair (Figure 9G).

DiscussionOther Section
NSUN1 is an RNA m5C methylase that mainly affects the expression or function of downstream molecules through epigenetic modifications. Studies have found that NSUN1 can regulate biological behaviors such as tumor cell growth and migration (12,13,18,19). At present, NSUN1 has been found to play a role in promoting cancer in 7 kinds of tumors, and high expression of NSUN1 predicts poor prognosis (8-13,20). So far, the research on NSUN1 is still in its infancy, with only few examples reported. Therefore, we combined scRNA-seq and Bulk RNA-seq data to conduct a comprehensive analysis of the molecular biological characteristics of NSUN1 based on the TCGA, Gene Expression Omnibus (GEO), and CPTAC databases, with the aim of revealing the role of NSUN1 in human cancers.
This study found that NSUN1 was mainly localized in the nucleus and was highly expressed in most tumor tissues. This study showed that NSUN1 was highly expressed in bladder urothelial carcinoma (BLCA), BRCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), CHOL, COAD, ESCA, glioblastoma multiforme (GBM), HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, STAD, uterine corpus endometrial carcinoma (UCEC). Furthermore, we found that NSUN1 overexpression generally predicted poor OS and/or DFS in patients with ACC, KICH, KIRC, KIRP, LAML, LGG, LIHC, MESO, SARC, SKCM, and UVM. These findings suggest that NSUN1 may be a valuable marker for early diagnosis and prognosis.
Predictive biomarkers in cancer patients are almost always based on the analysis of the entire biopsy sample, and their specific molecular mechanisms are still unclear, which affects the clinical application. The application of scRNA-seq technology has solved this problem to a certain extent. In this study, by analyzing scRNA-seq data, we found that in breast, liver, and stomach, the expression of NSUN1 was significantly higher in T cells, B cells, and DC cells than in other types of cells. This result suggests that NSUN1 may participate in various biological processes mainly by affecting the function of immune cells. In addition, this suggests that if NSUN1 becomes a molecular target for tumor therapy in the future, its main regulatory mechanism is to affect the functions of T cells and B cells to participate in molecular therapy.
The immune-related research results of this study found that NSUN1 negatively regulated the infiltration of immune cells in most tumors and participated in the regulation of the tumor immune microenvironment. NSUN1 inhibited the infiltration of DC cells, monocytes/macrophages and CD8+ T cells, especially in SKCM, STAD, TGCT. This result preliminarily confirmed our hypothesis: NSUN1 promotes the proliferation, invasion and metastasis of cancer cells by inhibiting the infiltration of immune cells in tumor tissues.
Phosphorylation is one of the most common post-translational modifications in proteins and is involved in a wide range of cellular activities including cell growth, differentiation and apoptosis (21,22). In cancer, the balance of activation and inactivation of many key kinases is delicately maintained through phosphorylation, and dysregulation of these processes results in disruption of signal transduction and metabolism. In this study, we found that the phosphorylation sites of NSUN1 were different in different tumors, but the phosphorylation level in tumors was higher than that in the corresponding normal tissues. This finding indicates that the phosphorylation of NSUN1 may be involved in the biological process of tumors as an important regulatory way. By targeting and regulating the phosphorylation of NSUN1, it may become a new approach for cancer treatment in the future. A limitation of this study is that all research findings are based on data analysis. The specific molecular mechanism still needs to be explored and discovered through basic research. The significance of this study is that it provides a new direction for future basic research.
ConclusionsOther Section
Four results of this study are very important. First, among all tumors with differential NSUN1 expression, NSUN1 expression was higher in tumor tissues. Second, in survival analysis, high NSUN1 expression predicted poor prognosis. Third, in different tumors, the phosphorylation modification level of tumor tissue was higher than that of normal tissue. Fourth, NSUN1 is associated with immune cell infiltration and may be involved in regulating the immune microenvironment. These findings indicate that NSUN1 plays a key role in the occurrence and development of tumors, and reflects its important value in the early diagnosis and prognosis of tumors.
AcknowledgmentsOther Section
We would like to thank Elvira Elizabeth (Shandong Foreign Language Vocational and Technical University) for her help in polishing our paper.
Funding: This work was supported by
FootnoteOther Section
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-66/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-66/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-66/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in compliance with the Helsinki Declaration (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Delaunay S, Pascual G, Feng B, et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 2022;607:593-603. [Crossref] [PubMed]
- Liao H, Gaur A, McConie H, et al. Human NOP2/NSUN1 regulates ribosome biogenesis through non-catalytic complex formation with box C/D snoRNPs. Nucleic Acids Res 2022;50:10695-716. [Crossref] [PubMed]
- Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol 2014;31:1-7. [Crossref] [PubMed]
- Kong W, Biswas A, Zhou D, et al. Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PloS Pathog 2020;16:e1008430. [Crossref] [PubMed]
- Yu J, Deshmukh H, Payton JE, et al. Array-based comparative genomic hybridization identifies CDK4 and FOXM1 alterations as independent predictors of survival in malignant peripheral nerve sheath tumor. Clin Cancer Res 2011;17:1924-34. [Crossref] [PubMed]
- Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 2013;5:237-47. [Crossref] [PubMed]
- Heissenberger C, Rollins JA, Krammer TL, et al. The ribosomal RNA m(5)C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans. Elife 2020;9:e56205. [Crossref] [PubMed]
- Fonagy A, Swiderski C, Ostrovsky AM, et al. Effect of nucleolar P120 expression level on the proliferation capacity of breast cancer cells. Cancer Res 1994;54:1859-64. [PubMed]
- Bi J, Huang Y, Liu Y. Effect of NOP2 knockdown on colon cancer cell proliferation, migration, and invasion. Transl Cancer Res 2019;8:2274-83. [Crossref] [PubMed]
- Feng J, Zhang J, Li Y, et al. Upregulated expression of NOP2 predicts worse prognosis of gastric adenocarcinoma by promoting tumor growth. Saudi J Gastroenterol 2022;28:369-77. [Crossref] [PubMed]
- Sun F, Wu K, Yao Z, et al. Long Noncoding RNA PVT1 Promotes Prostate Cancer Metastasis by Increasing NOP2 Expression via Targeting Tumor Suppressor MicroRNAs. Onco Targets Ther 2020;13:6755-65. [Crossref] [PubMed]
- Sun F, Wu K, Yao Z, et al. Long noncoding RNA LINC00963 induces NOP2 expression by sponging tumor suppressor miR-542-3p to promote metastasis in prostate cancer. Aging (Albany NY) 2020;12:11500-16. [Crossref] [PubMed]
- Wang G, Qu F, Liu S, et al. Nucleolar protein NOP2 could serve as a potential prognostic predictor for clear cell renal cell carcinoma. Bioengineered 2021;12:4841-55. [Crossref] [PubMed]
- Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 2014;60:1278-90. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Saijo Y, Sato G, Usui K, et al. Expression of nucleolar protein p120 predicts poor prognosis in patients with stage I lung adenocarcinoma. Ann Oncol 2001;12:1121-5. [Crossref] [PubMed]
- Saijo Y, Perlaky L, Valdez BC, et al. The effect of antisense p120 construct on p120 expression and cell proliferation in human breast cancer MCF-7 cells. Cancer Lett 1993;68:95-104. [Crossref] [PubMed]
- Gao Y, Wang Z, Zhu Y, et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci 2019;110:3510-9. [Crossref] [PubMed]
- Bucci M. Functional phosphorylation. Nat Chem Biol 2018;14:525. [Crossref] [PubMed]
- Kholodenko BN, Okada M. Reengineering protein-phosphorylation switches. Science 2021;373:25-6. [Crossref] [PubMed]


