
Dihydroorotate dehydrogenase promotes cell proliferation and suppresses cell death in esophageal squamous cell carcinoma and colorectal carcinoma
Highlight box
Key findings
• Dihydroorotate dehydrogenase (DHODH) was overexpressed in esophageal carcinoma (ESCA), colon adenocarcinoma (COAD), and rectum adenocarcinoma (READ). Silence and inactivation of DHODH inhibited the proliferation and induced cell death of esophageal squamous cell carcinoma (ESCC) and colorectal cancer (CRC) cells.
What is known and what is new?
• DHODH is newly identified as an anti-ferroptosis molecule independent from the well-known GPX4 and AIFM2.
• Silence and inactivation of DHODH inhibited the proliferation and induced cell death of ESCC and CRC cells.
What is the implication, and what should change now?
• Targeting DHODH-induced ferroptosis will probably be used to treat DHODH-overexpressed cancers in future.
Introduction
As a newly-defined form of cell death, ferroptosis has the main characteristic of iron-dependent lipid peroxidation (1). Ferroptosis is regarded as a promising natural anti-tumor therapeutic method (2). AIFM2 and GPX4 have been identified as the 2 major ferroptosis defense systems (3-6). Dihydroorotate dehydrogenase (DHODH) is a newly discovered anti-ferroptosis molecule independent from GPX4 and AIFM2, the inactivation of which causes enhanced mitochondrial peroxidation and ferroptosis in GPX4-lowly-expressed cancer cells (7). However, the expression pattern, prognostic role, and especially the ferroptosis-related functional role of DHODH in pan-cancer are still largely unclear.
DHODH shows important roles during de novo pyrimidine biosynthesis, and its suppression induces apoptosis and differentiation of acute myeloid leukemia (AML) cells. Its inhibitor MEDS433 could synergically induce the metabolic lethality of AML cells with dipyridamole which is an inhibitor of the pyrimidine salvage pathway (8). In oral squamous cell carcinoma (OSCC), DHODH was found to be overexpressed and its elevated expression was significantly correlated with patients’ unfavorable prognosis. Besides, inactivation of DHODH by either inhibitor or short hairpin RNA (shRNA) was shown to impede OSCC cell proliferation in vitro and tumor growth in vivo (9). In esophageal squamous cell carcinoma (ESCC), DHODH directly bound to the NH2 terminal of β-catenin, inhibited its protein degradation, and promoted its nuclear accumulation, then finally activated the β-catenin signaling pathway (10). DHODH together with PCK1 was found to promote the liver metastatic colonization and hypoxic growth of colorectal cancer (CRC) via enhancing nucleotide synthesis (11). Recent studies have reported that blocking SOD2 could enhance ferroptosis and augment radiosensitivity of nasopharyngeal carcinoma via regulating DHODH activity, and that suppression of DHODH synergistically improved therapeutic effect with cisplatin in cervical cancer via inducting ferroptosis (12,13). However, the roles of DHODH in the ferroptosis of ESCC and CRC cells are still unclear.
In the present study, we analyzed the expression level and prognostic value of DHODH in cancers, and importantly explored the roles of DHODH in the proliferation and cell death of ESCC and CRC cells. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-136/rc).
Methods
Patients and tissues
A total of 20 surgical resected ESCC tissues were collected at First People’s Hospital of Yunnan Province and Affiliated Hospital of Kunming University of Science and Technology (Kunming, China). All participants had not received treatment before surgery and provided informed consent. The Medical Ethics Committee of Kunming University of Science and Technology approved this study (No. KMUST-MEC-123). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell lines, transfection, and reagents
KYSE450 and KYSE510 ESCC and SW620 CRC cell lines were cultured in Roswell Park Memorial Institute (RPMI; Buffalo, NY, USA) 1640 cell culture medium [containing 10% fetal bovine serum (FBS), 100 mg/mL streptomycin, and 100 U/mL penicillin] under the conditions of 37 °C and 5% CO2. Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was applied for cell transfection.
DHODH and negative control small interfering RNAs (siRNAs) were produced by GenePharma (Shanghai, China). Leflunomide (LEF), Z-VAD-FMK, necrostatin-1, liproxstatin-1, and ferrostatin-1 were provided by Selleck (Houston, TX, USA).
Total RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Previous methods were applied to extract total RNA and perform qRT-PCR assay (14). Details of the primers used are provided in Table S1.
Western blot assay
A previously described method was applied for western blot analysis (14). The antibodies were as follows: DHODH antibody (14877-1-AP; Proteintech, Wuhan, China), GPX4 antibody (ab125066; Abcam, Cambridge, MA, USA), SLC7A11 antibody (ab175186; Abcam), and GAPDH antibody (60004-1-IG; Proteintech).
Cell viability, proliferation, colony formation, and cellular adenosine triphosphate (ATP) assays
Previous methods were applied to detect cell viability and abilities of cell proliferation and colony formation (14). Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Tokyo, Japan) was harnessed to measure cell viability and proliferation ability. In colony formation, clones were stained by crystal violet (Sigma Aldrich, St. Louis, MO, USA) and further visualized and quantitated. Cellular ATP levels were detected using a Beyotime enhanced ATP assay kit (Beyotime, Shanghai, China).
Messenger RNA (mRNA) sequencing
RNA extraction and mRNA sequencing were performed by BGI Genomics Co., Ltd. (Shenzhen, China). Total RNA was prepared using TRIzol™ reagent (Thermo Fisher). An Agilent 2100 Bioanalyzer system (Santa Clara, CA, USA) was harnessed to measure RNA quantity and quality using an RNA Nano 6000 Kit. The mRNAs from samples were sequenced by DNBSEQ platform (MGI Tech., Shenzhen, China).
Differentially expressed genes (DEGs) were identified through comparing normalized reads count between DHODH-silenced and control cells with absolute log2 [fold change (FC)]>1.0 and P<0.05.
Databases analyses
The Kaplan-Meier Plotter (15), Tumor Immune Estimation Resource (TIMER) (16-18), Gene Expression Profiling Interactive Analysis (GEPIA) (19) databases were applied to analyze expression pattern, prognostic role, and correlations between indicated genes in pan-cancer.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses
KEGG pathway enrichment and GO analyses were carried out on the Database for Annotation, Visualization and Integrated Discovery (DAVID) platform (20,21).
Statistical analyses
Statistical analysis was carried out through GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). Data were shown as mean ± standard deviation (SD) and analyzed by analysis of variance (ANOVA) and Student’s t-test. Kaplan-Meier method was used to estimate overall survival (OS) time. Statistical significance was considered when P<0.05.
Results
Pan-cancer analysis of DHODH expression pattern and its prognostic role
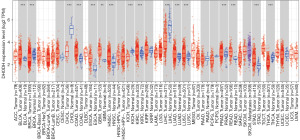
Analysis of the TIMER database revealed that DHODH was overexpressed in many types of cancers such as esophageal carcinoma (ESCA), colon adenocarcinoma (COAD), and rectum adenocarcinoma (READ). Using the GEPIA database, DHODH expression was found to be lower in liver hepatocellular carcinoma (LIHC) and higher in diffuse large B-cell lymphoma (DLBC), glioblastoma (GBM), thymoma (THYM), and lower grade glioma (LGG) comparing with corresponding normal tissues (Figure S1A,S1B).
Pan-cancer prognosis analysis of DHODH was achieved via analyzing the Kaplan-Meier Plotter, GEPIA, and TIMER databases. High expression of DHODH was correlated with improved OS and relapse-free survival (RFS) of kidney renal papillary cell carcinoma (KIRP) and lung adenocarcinoma (LUAD) patients, and associated with shortened OS as well as RFS of sarcoma (SARC) patients (Figure S2A-S2F). In the TIMER database, DHODH high expression indicated poor prognosis in SARC but had no prognostic value in KIRP (Figure S2G,S2H). In the Kaplan-Meier Plotter database, DHODH high expression was positively linked with good OS in liver cancer, READ, THYM, and uterine corpus endometrial carcinoma (UCEC), and poor RFS of cervical squamous cell carcinoma (CSCC) and stomach adenocarcinoma (STAD) patients, and improved RFS in thyroid cancer (THCA) (Figure S2I-S2O). In the GEPIA database, correlations between DHODH high expression and good OS in KIRC or poor disease-free survival (DFS) in pancreatic adenocarcinoma (PAAD) or improved DFS in LIHC were identified (Figure S3A-S3D).
Interestingly, after analyzing the TIMER database, data showed that DHODH mRNA expression was higher in TP53-mutated bladder urothelial carcinoma (BLCA), ESCA, human papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSC), luminal-A breast invasive carcinoma (BRCA), and lower in TP53-mutated uterine carcinosarcoma (UCS) and LIHC (Figure S4A-S4F), and also higher in PTEN-mutated KIRP, prostate adenocarcinoma (PRAD), UCEC, and BLCA (Figure S5A-S5D).
Considering that DHODH was overexpressed in GBM and under-expressed in LIHC both in GEPIA and TIMER databases, GO and KEGG analyses were further performed based on DEGs both in GBM [n=500, Pearson correlation coefficient (PCC): 0.79–0.57] and LIHC (n=100, PCC: 0.73–0.33) by DAVID. In GBM, biological process (BP) enrichment analysis indicated that DEGs were correlated with transcription, mRNA splicing, and so on (Figure S6A and Table S2); cellular component (CC) enrichment analysis indicated that DEGs were correlated with nucleoplasm, nucleus, and so on (Figure S6B and Table S3); molecular function (MF) enrichment analysis indicated that DEGs were correlated with nucleic acid binding, poly (A) RNA binding, and so on (Figure S6C and Table S4); KEGG enrichment analysis indicated that DEGs were correlated with spliceosome, mRNA surveillance pathway, and so on (Figure S6D and Table S5). In LIHC, BP were enriched in acute-phase response and so on (Figure S6E and Table S6); CC were enriched in blood microparticle and so on (Figure S6F and Table S7); MF were enriched in serine-type endopeptidase activity and so on (Figure S6G and Table S8); KEGG analysis indicated that DEGs were enriched in complement and coagulation cascades and so on (Figure S6H and Table S9).
Then, we evaluated the associations between DHODH and ferroptosis-associated genes including GPX4, SLC7A11, and AIFM2 using the GEPIA database, and positive correlation (P<0.05 and R≥0.2) suggested that DHODH might participate in ferroptosis by cross-linking with GPX4/SLC7A11/AIFM2. Positive correlations between DHODH and GPX4 were detected in ESCA (P=0.002, R=0.230) together with kidney chromophobe (KICH; P=0.003, R=0.360), and between DHODH and SLC7A11 in many types of cancers such as GBM and lung squamous cell carcinoma (LUSC), Table S10), and between DHODH and AIFM2 in CESC, ESCA, and so on (Table S10).
DHODH was overexpressed in ESCC and CRC
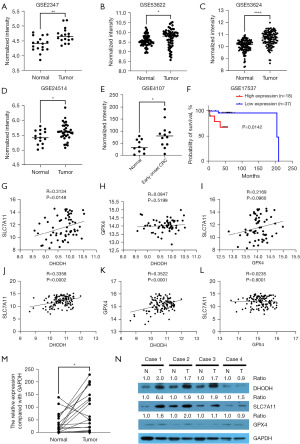
Figure 1 shows that DHODH was overexpressed in ESCA, COAD, and READ in mRNA level. Overexpression of DHODH in ESCC and CRC was further validated via analyzing ESCC datasets (GSE20347, GSE53622, and GSE53624; Figure 2A-2C) and CRC datasets (GSE24514 and GSE4107; Figure 2D,2E). Importantly, DHODH high expression was markedly associated with unfavorable outcome in CRC (Figure 2F). Expression of DHODH was positively correlated with SLC7A11 mRNA level both in GSE53622 and GSE53624 datasets, and correlation between expressions of DHODH and GPX4 were only identified in GSE53624 dataset, and there was no correlation between expressions of SLC7A11 and GPX4 in 2 datasets (Figure 2G-2L). The qRT-PCR assay verified higher expression of DHODH in ESCC samples comparing to normal tissues (Figure 2M, n=20). Using western blot, protein expressions of DHODH, SLC7A11, and GPX4 in ESCC tissues were detected, moreover DHODH, SLC7A11, and GPX4 were overexpressed in 3, 4, and 2 ESCC samples respectively (ratio ≥1.5; Figure 2N).
Silence or inactivation of DHODH inhibited the proliferation and induced the cell death in ESCC and CRC cells
Knockdown of DHODH remarkably suppressed ESCC (KYSE510 and KYSE450) and CRC (SW620) cells’ proliferation (Figure 3A-3D). Colony formation abilities of DHODH-silenced KYSE510 and KYSE450 cells were descended (Figure 3E). Interestingly, knockdown of DHODH decreased the cellular ATP levels in KYSE510, KYSE450, and SW620 cells (Figure 3F-3H).

LEF, a clinically approved DHODH inhibitor, was harnessed to evaluate the influences of blocking DHODH on the phenotypes of cancer cells. The IC50 of LEF in KYSE510, KYSE450, and SW620 cells was 108.2 µM, 124.8 µM, and 173.9 µM, respectively (Figure 4A-4C). LEF significantly reduced viabilities of KYSE510, KYSE450, and SW620 cells (Figure 4D). Colony formation assay showed that LEF treatment markedly reduced the colony formation abilities of both ESCC cells and CRC cells (Figure 4E,4F). LEF also significantly reduced the cellular ATP levels in KYSE510, KYSE450, and SW620 cells (Figure 4G). Importantly, the inhibitory effects on ESCC cell viabilities of LEF could be partially recovered by the inhibitor of apoptosis Z-VAD-FMK, moreover the inhibition on cell viability of CRC cells caused by LEF could be partially impeded by ferroptosis inhibitors (Lip-1, Liproxstatin-1; Fer-1, Ferrostatin-1) and necroptosis inhibitor (Nec-1, Necrostatin-1; Figure 4H-4J). These results suggested that DHODH promoted the proliferation and suppressed the cell death both in ESCC and CRC.

Identification of downstream target genes and pathways of DHODH in ESCC
DEGs after DHODH knockdown were screened by using RNA-seq method. After DHODH knockdown, 27 genes were downregulated and 14 genes were upregulated both in KYSE510 and KYSE450 cells (Figure 5A-5C and Table 1). The top 5 downregulated genes were U2AF1, SULT1A4, EIF3CL, DHODH, and TXNDC12, and the top 5 upregulated genes were H4C12, SDR16C5, GDF15, TMED7-TICAM2, and GPR75. The CC of extracellular space was enriched both in KYSE510 and KYSE450 cells (Figure 5D,5E). There was no common BP both in KYSE510 and KYSE450 (Figure 5F,5G). The MF of growth factor activity was enriched both in KYSE510 and KYSE450 cells (Figure 6A,6B). The enriched pathways were rheumatoid arthritis, salmonella infection, cytokine-cytokine receptor interaction, pertussis, and nuclear factor-κB (NF-κB) (Figure 6C,6D). In qRT-PCR validation experiment, silence of DHODH downregulated TXNDC, USE1, and U2AF1 and upregulated HMOX1 and PTPRB both in KYSE510 and KYSE450 cells (Figure 6E,6F). Besides that, LEF treatment also significantly decreased the mRNA levels of DHODH, TXNDC12, USE1, and U2AF1 both in KYSE510 and KYSE 450 cells (Figure 6G,6H).

Table 1
| No. | Gene ID | Gene symbol | Regulation | KYSE450 | KYSE510 | |||
|---|---|---|---|---|---|---|---|---|
| log2 (si2/NC) | FDR (si2/NC) | log2 (si2/NC) | FDR (si2/NC) | |||||
| 1 | 8362 | H4C12 | Up | 10.013 | 1.60E−04 | 9.392 | 6.32E−04 | |
| 2 | 195814 | SDR16C5 | Up | 2.921 | 4.27E−04 | 2.772 | 3.36E−28 | |
| 3 | 9518 | GDF15 | Up | 1.881 | 1.67E−87 | 1.843 | 0 | |
| 4 | 100302736 | TMED7-TICAM2 | Up | 1.846 | 1.50E−28 | 1.197 | 7.29E−21 | |
| 5 | 10936 | GPR75 | Up | 1.828 | 2.88E−05 | 1.143 | 2.60E−05 | |
| 6 | 102800317 | TPTEP2-CSNK1E | Up | 1.732 | 2.71E−04 | 1.874 | 1.44E−06 | |
| 7 | 55586 | MIOX | Up | 1.598 | 3.48E−04 | 3.949 | 4.50E−12 | |
| 8 | 3162 | HMOX1 | Up | 1.448 | 2.30E−08 | 2.949 | 0 | |
| 9 | 5328 | PLAU | Up | 1.291 | 1.17E−124 | 1.332 | 5.49E−60 | |
| 10 | 1839 | HBEGF | Up | 1.190 | 2.27E−17 | 2.852 | 0 | |
| 11 | 5787 | PTPRB | Up | 1.142 | 2.49E−06 | 1.788 | 6.04E−05 | |
| 12 | 79705 | LRRK1 | Up | 1.099 | 0 | 1.136 | 2.39E−178 | |
| 13 | 1073 | CFL2 | Up | 1.078 | 3.77E−16 | 1.054 | 1.86E−40 | |
| 14 | 118788 | PIK3AP1 | Up | 1.011 | 3.26E−15 | 2.954 | 1.66E−07 | |
| 15 | 7307 | U2AF1 | Down | −4.214 | 8.80E−109 | −4.363 | 1.37E−218 | |
| 16 | 445329 | SULT1A4 | Down | −2.324 | 2.43E−72 | −10.329 | 9.97E−65 | |
| 17 | 728689 | EIF3CL | Down | −1.735 | 5.30E−252 | −1.411 | 2.45E−35 | |
| 18 | 1723 | DHODH | Down | −1.634 | 1.98E−29 | −2.548 | 6.52E−35 | |
| 19 | 51060 | TXNDC12 | Down | −1.414 | 2.33E−80 | −1.205 | 1.29E−84 | |
| 20 | 65989 | DLK2 | Down | −1.374 | 1.05E−18 | −1.306 | 2.37E−27 | |
| 21 | 388963 | C2orf81 | Down | −1.336 | 3.85E−12 | −1.724 | 7.76E−04 | |
| 22 | 201266 | SLC39A11 | Down | −1.333 | 5.55E−20 | −1.569 | 1.38E−12 | |
| 23 | 1397 | CRIP2 | Down | −1.293 | 5.71E−08 | −1.167 | 3.75E−11 | |
| 24 | 5275 | SERPINB13 | Down | −1.241 | 1.03E−17 | −1.201 | 6.52E−39 | |
| 25 | 55646 | LYAR | Down | −1.209 | 1.90E−41 | −1.698 | 1.01E−85 | |
| 26 | 1207 | CLNS1A | Down | −1.197 | 3.84E−217 | −1.934 | 4.40E−242 | |
| 27 | 112495 | GTF3C6 | Down | −1.184 | 9.64E−11 | −1.518 | 1.75E−44 | |
| 28 | 114971 | PTPMT1 | Down | −1.160 | 4.78E−38 | −1.419 | 9.39E−83 | |
| 29 | 51529 | ANAPC11 | Down | −1.141 | 1.99E−25 | −1.044 | 2.33E−21 | |
| 30 | 55052 | MRPL20 | Down | −1.135 | 3.19E−22 | −1.130 | 1.52E−42 | |
| 31 | 10994 | ILVBL | Down | −1.133 | 3.02E−35 | −1.335 | 1.11E−37 | |
| 32 | 79077 | DCTPP1 | Down | −1.111 | 2.83E−46 | −1.562 | 2.88E−72 | |
| 33 | 4059 | BCAM | Down | −1.094 | 1.21E−26 | −1.263 | 3.94E−34 | |
| 34 | 2700 | GJA3 | Down | −1.063 | 1.43E−09 | −1.190 | 8.21E−04 | |
| 35 | 285148 | IAH1 | Down | −1.062 | 2.93E−29 | −2.027 | 2.97E−13 | |
| 36 | 55850 | USE1 | Down | −1.037 | 1.34E−06 | −1.035 | 3.94E−12 | |
| 37 | 60558 | GUF1 | Down | −1.023 | 2.62E−37 | −1.493 | 3.65E−59 | |
| 38 | 54892 | NCAPG2 | Down | −1.020 | 1.16E−21 | −1.624 | 7.03E−98 | |
| 39 | 151188 | ARL6IP6 | Down | −1.008 | 5.69E−18 | −1.256 | 7.80E−17 | |
| 40 | 84992 | PIGY | Down | −1.006 | 2.84E−04 | −1.681 | 4.68E−20 | |
| 41 | 5533 | PPP3CC | Down | −1.005 | 2.37E−06 | −1.690 | 4.75E−18 | |
DEGs, differentially expressed genes; DHODH, dihydroorotate dehydrogenase; NC, negative control; FDR, false discovery rate.

Discussion
DHODH plays an important role in de novo pyrimidine synthesis and is a flavin-dependent mitochondrial enzyme (22). The synthesis of pyrimidine nucleotides consists of 2 pathways including the de novo pathway and salvage pathway. In rapidly proliferative cells, pyrimidine nucleotides are mainly synthesized by the de novo pathway, and in fully differentiated cells, pyrimidines nucleotides are mainly obtained by the salvage pathway (23,24). DHODH is located in mitochondria inner membrane because the N-terminal of human DHODH protein contains the mitochondrial-targeting pre-sequences (25). However, recent research has reported that DHODH functioned as a new anti-ferroptosis gene independent from GPX4 and AIFM2 via affecting the mitochondrial lipid peroxidation (7). Delivering multi-siRNAs targeting GPX4 and DHODH using exosomes could augment sorafenib-induced ferroptosis of hepatocellular carcinoma cells (26). Manganese treatment could inhibit DHODH activity via regulating type-I interferons (IFNs) and eventually lead to ferroptosis of cancer cells (27). However, its roles and mechanisms in the ferroptosis of many other cancers, especially ESCC and CRC, are still largely unclear, and whether its anti-ferroptosis function is associated with its roles in de novo pyrimidine synthesis or its location in mitochondria inner membrane is still unknown.
Ferroptosis has been shown to participate in the development and therapeutic responses of many kinds of tumors, and damage of ferroptosis could induce inflammation-linked immunosuppression in the microenvironment of cancer and promote cancer growth (1). In the GEPIA and TIMER databases, DHODH was overexpressed in GBM and under-expressed in LIHC. In high grade gliomas and GBM cell lines, DHODH was elevated, and in particular, inhibition of DHODH suppresses the proliferation of GBM cells (28). Blockade of DHODH activity could inhibit the pyrimidine synthesis and the tumorigenic capacity of GBM stem cells (29). However, whether the suppression on proliferation of GBM cells caused by DHODH inhibition is associated with ferroptosis still needed to be explored. The role and regulatory mechanism of DHODH downregulation in LIHC are also needed to be studied.
In this current study, DHODH overexpression was found to be linked with improved OS in KIRP and decreased OS in SARC. A previous study reported that LEF which was an inhibitor of DHODH significantly impeded the proliferation of renal carcinoma cells and tumor growth in an animal model through suppressing the canonical Wnt/β-catenin signaling pathway (30). Whether the dysregulation of DHODH is linked with ferroptosis of renal carcinoma cells and the role of DHODH in SARC need to be studied in the future.
The current data indicated that the mRNA expression level of DHODH was higher in TP53-mutated BLCA, ESCA, HPV-negative HNSC, and luminal-A BRCA patients and in PTEN-mutated KIRP, PRAD, UCEC, and BLCA patients. These findings suggest that there might be regulatory connections between DHODH expression and mutations of TP53 and PTEN. Tenovins was reported to activate p53 and kill cancer cells by inhibiting DHODH activity and blocking uridine transport into cells (31). The inhibitors of DHODH, tetrahydroindazoles, could suppress cancer cell growth and viability by activating p53 and its downstream genes (32). DHODH inhibitors could enhance the p53 synthesis and promote cancer cell death through blocking p53 degradation (33). Inhibition of DHODH could also reduce ATP level and increase the reactive oxygen species (ROS) level of breast cancer cells via upregulating p53, p65, and STAT6, and finally inhibit the proliferation of cancer cells (34). All these findings highlight DHODH as an upstream regulatory gene in the p53 signaling pathway, however, the expression level of DHODH in p53 mutant cancer cells is still unclear and needs to be evaluated. A previous study reported that mutations in PTEN generated different dihydroorotase (a rate-limiting step enzyme in pyrimidine synthesis) phosphorylation patterns and further activated carbon influx by regulating pyrimidine synthesis in GBM stem cells (29). Nevertheless, the regulatory mechanism underlying mutation of PTEN and DHODH expression or activity is still unknown.
Recently, a study reported that combination of the DHODH inhibitor LEF with RGX-202 (an inhibitor of SLC6A8) suppressed CRC growth in animal models (35). Importantly, our results further showed that knockdown and DHODH inhibitor LEF suppressed the cell proliferation, colony formation, and cellular ATP levels in ESCC and CRC cells. Interestingly, the apoptosis inhibitor partially rescued LEF-induced cell death of ESCC cells; moreover, ferroptosis and necroptosis inhibitors partially rescued LEF-induced cell death of CRC cells, respectively. These results suggest that LEF induced apoptosis of ESCC cells and led to ferroptosis and necroptosis of CRC cells. Up to now, the roles and mechanisms of DHODH in cell death, especially the necroptosis and ferroptosis of tumor cells, are almost completely unknown, thus, future studies should aim to explore the regulatory roles of DHODH during ferroptosis and necroptosis of cancer cells.
Conclusions
Taken together, our results indicate that DHODH is dysregulated in many kinds of tumors including ESCA, COAD, and READ. Importantly, DHODH was shown to suppress apoptosis in ESCC and inhibit ferroptosis and necroptosis in CRC. Whether a therapeutic method of targeting DHODH-induced ferroptosis could be used to treat DHODH-overexpressed cancers needs to be explored in future.
Acknowledgments
Funding: This study was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-136/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-136/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-136/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-136/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Medical Ethics Committee of Kunming University of Science and Technology approved this study (No. KMUST-MEC-123) and informed consent was provided by all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 2021;18:280-96. [Crossref] [PubMed]
- Wang H, Cheng Y, Mao C, et al. Emerging mechanisms and targeted therapy of ferroptosis in cancer. Mol Ther 2021;29:2185-208. [Crossref] [PubMed]
- Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 2014;16:1180-91. [Crossref] [PubMed]
- Yang WS. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. [Crossref] [PubMed]
- Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019;575:688-92. [Crossref] [PubMed]
- Doll S, Freitas FP, Shah R, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 2019;575:693-8. [Crossref] [PubMed]
- Mao C, Liu X, Zhang Y, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 2021;593:586-90. [Crossref] [PubMed]
- Gaidano V, Houshmand M, Vitale N, et al. The Synergism between DHODH Inhibitors and Dipyridamole Leads to Metabolic Lethality in Acute Myeloid Leukemia. Cancers (Basel) 2021;13:1003. [Crossref] [PubMed]
- Qiu X, Jiang S, Xiao Y, et al. SOX2-dependent expression of dihydroorotate dehydrogenase regulates oral squamous cell carcinoma cell proliferation. Int J Oral Sci 2021;13:3. [Crossref] [PubMed]
- Qian Y, Liang X, Kong P, et al. Elevated DHODH expression promotes cell proliferation via stabilizing β-catenin in esophageal squamous cell carcinoma. Cell Death Dis 2020;11:862. [Crossref] [PubMed]
- Yamaguchi N, Weinberg EM, Nguyen A, et al. PCK1 and DHODH drive colorectal cancer liver metastatic colonization and hypoxic growth by promoting nucleotide synthesis. Elife 2019;8:e52135. [Crossref] [PubMed]
- Jiang M, Song Y, Liu H, et al. DHODH Inhibition Exerts Synergistic Therapeutic Effect with Cisplatin to Induce Ferroptosis in Cervical Cancer through Regulating mTOR Pathway. Cancers (Basel) 2023;15:546. [Crossref] [PubMed]
- Amos A, Jiang N, Zong D, et al. Depletion of SOD2 enhances nasopharyngeal carcinoma cell radiosensitivity via ferroptosis induction modulated by DHODH inhibition. BMC Cancer 2023;23:117. [Crossref] [PubMed]
- Shi ZZ, Wang WJ, Chen YX, et al. The miR-1224-5p/TNS4/EGFR axis inhibits tumour progression in oesophageal squamous cell carcinoma. Cell Death Dis 2020;11:597. [Crossref] [PubMed]
- Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11:6047. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Zhou Y, Tao L, Zhou X, et al. DHODH and cancer: promising prospects to be explored. Cancer Metab 2021;9:22. [Crossref] [PubMed]
- Löffler M, Fairbanks LD, Zameitat E, et al. Pyrimidine pathways in health and disease. Trends Mol Med 2005;11:430-7. [Crossref] [PubMed]
- Mathur D, Stratikopoulos E, Ozturk S, et al. PTEN Regulates Glutamine Flux to Pyrimidine Synthesis and Sensitivity to Dihydroorotate Dehydrogenase Inhibition. Cancer Discov 2017;7:380-90. [Crossref] [PubMed]
- Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem 2004;279:33035-8. [Crossref] [PubMed]
- Li X, Yu Q, Zhao R, et al. Designer Exosomes for Targeted Delivery of a Novel Therapeutic Cargo to Enhance Sorafenib-Mediated Ferroptosis in Hepatocellular Carcinoma. Front Oncol 2022;12:898156. [Crossref] [PubMed]
- Zhang S, Kang L, Dai X, et al. Manganese induces tumor cell ferroptosis through type-I IFN dependent inhibition of mitochondrial dihydroorotate dehydrogenase. Free Radic Biol Med 2022;193:202-12. [Crossref] [PubMed]
- Lafita-Navarro MC, Venkateswaran N, Kilgore JA, et al. Inhibition of the de novo pyrimidine biosynthesis pathway limits ribosomal RNA transcription causing nucleolar stress in glioblastoma cells. PLoS Genet 2020;16:e1009117. [Crossref] [PubMed]
- Wang X, Yang K, Wu Q, et al. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci Transl Med 2019;11:eaau4972. [Crossref] [PubMed]
- Chen Y, Huang Q, Zhou H, et al. Inhibition of canonical WNT/beta-catenin signaling is involved in leflunomide (LEF)-mediated cytotoxic effects on renal carcinoma cells. Oncotarget 2016;7:50401-16. [Crossref] [PubMed]
- Ladds MJGW, Popova G, Pastor-Fernández A, et al. Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology. J Biol Chem 2020;295:17935-49. [Crossref] [PubMed]
- Popova G, Ladds MJGW, Johansson L, et al. Optimization of Tetrahydroindazoles as Inhibitors of Human Dihydroorotate Dehydrogenase and Evaluation of Their Activity and In Vitro Metabolic Stability. J Med Chem 2020;63:3915-34. [Crossref] [PubMed]
- Ladds MJGW, van Leeuwen IMM, Drummond CJ, et al. A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage. Nat Commun 2018;9:1107. [Crossref] [PubMed]
- Mohamad Fairus AK, Choudhary B, Hosahalli S, et al. Dihydroorotate dehydrogenase (DHODH) inhibitors affect ATP depletion, endogenous ROS and mediate S-phase arrest in breast cancer cells. Biochimie 2017;135:154-63. [Crossref] [PubMed]
- Kurth I, Yamaguchi N, Andreu-Agullo C, et al. Therapeutic targeting of SLC6A8 creatine transporter suppresses colon cancer progression and modulates human creatine levels. Sci Adv 2021;7:eabi7511. [Crossref] [PubMed]


