
Ubenimex suppresses glycolysis mediated by CD13/Hedgehog signaling to enhance the effect of cisplatin in liver cancer
Highlight box
Key findings
• Ubenimex inhibits glycolysis by targeting the CD13/Hh pathway, thus playing an anti-tumor role together with CDDP.
What is known and what is new?
• As a CD13 inhibitor, ubenimex can enhance the efficacy of chemotherapeutic drugs.
• In this study, ubenimex was also shown to suppress glycolysis meditated by CD13/Hh signaling to enhance the effect of CDDP in liver cancer.
What is the implication, and what should change now?
• This study clarified a new regulatory mechanism between CD13 molecule and the therapeutic effect of CDDP, thus providing theoretical basis for enhancing the anti-tumor effect of CDDP in liver cancer.
Introduction
Liver cancer as a common malignant tumor ranks third among all cancer-related deaths worldwide because of its high invasion and metastasis rate (1,2). Hepatocellular carcinoma (HCC) accounts for 90% of primary liver cancer (3). Despite the application of a multitude of available therapies, the 5-year survival rate and prognosis of patients with liver cancer are still unfavorable (4). cis-Diamminedichloroplatinum (II) (cisplatin, CDDP), which is one of the conventional chemotherapeutic drugs, is indispensable in the comprehensive treatment of unresectable or metastatic liver cancer (5,6). Unfortunately, severe toxic effects and chemoresistance greatly subside the clinical efficacy of CDDP (7). Thus, novel therapeutic regimens are required to improve the antitumor therapeutic outcomes of CDDP.
Even in the presence of sufficient oxygen, the majority of cancer cells metabolize glucose mainly through glycolysis rather than oxidative phosphorylation, which provides sufficient energy and materials for the rapid proliferation of tumor cells (8). As a hallmark of cancer, aerobic glycolysis is considered to be essential for the proliferation, invasion and metastasis, angiogenesis, and immune evasion of cancer cells (9,10). Glycolytic transcriptional regulators and glycolysis-related proteins in cancers are significantly related to poor prognosis in many types of cancers, including liver cancer (11-13). High tumor glycolytic activity is associated with inferior overall survival in the pan-cancer patients (14). Glycolysis-associated multi-omics prognostic model (GMPM) is confirmed as an independent risk factor for the prognosis of patients with HCC (15). By stratifying the patients with HCC with glycolysis-associated genes, GMPM may provide a more efficient way of screening high-risk patients with HCC associated with poor prognosis (15). The key molecules of glycolysis, including hexokinase 2 (HK2), pyruvate kinase M2 (PKM2), and glucose transporter 1 (GLUT1), are highly expressed in liver cancer, promoting cell proliferation and metastasis (16-20).
Hedgehog (Hh) signaling is vital for controlling tumor growth, cell proliferation, and differentiation during embryonic development and carcinogenesis (21). Following binding of Hh ligands to the membrane receptor patched homolog 1 (PTCH1), the signal transduction is initiated and Smoothened (SMO) is no longer repressed by PTCH1. Following activation, SMO releases glioma-associated oncogene (GLI) from its inhibitors, promoting the transcription of the downstream target genes (22,23). The Hh pathway is abnormally activated in multiple types of cancers, including liver cancer, and promotes tumor invasion, metastasis, and multidrug resistance (24-26). Hh is the most highly expressed ligand in liver cancer (27). Growing evidence indicates that the Hh pathway is closely associated with aerobic glycolysis in a variety of diseases (28-33). However, the underlying molecular mechanism of Hh signaling-mediated aerobic glycolysis in liver cancer has not been fully explained.
Cluster of differentiation 13 (CD13), also known as aminopeptidase N (ANPEP), is an important zinc-dependent type II metalloexopeptidase (34). CD13 has been shown to be overexpressed in a variety of cancer types including liver cancer; it promotes tumor angiogenesis, proliferation, invasion, and metastasis through the interaction between its enzyme activity and signal function (34-36). As the only marketed CD13 inhibitor, ubenimex has been observed to exert synergistic antitumor effects with chemotherapeutic drugs by suppressing the self-renewal of cancer stem cells, inhibiting cancer cell autophagy, promoting chemotherapy-induced apoptosis, and decreasing the expression of membrane transport proteins (37-40). To the best of our knowledge, the adjuvant chemotherapy effect of ubenimex has seldom been evaluated from the viewpoint of Hh signaling-dependent glycolytic regulation.
Although certain experiments have shown that ubenimex can synergistically potentiate the antitumor effect of chemotherapy, the mechanisms differ. The present study aimed to provide further understanding on the synergistic effect of ubenimex with CDDP in liver cancer, and to investigate the possible associated molecular events. The data demonstrated that ubenimex can regulate glycolysis by targeting the CD13/Hh pathway, so as to exert its adjuvant chemotherapeutic effect. Collectively, the emerging regulatory mechanisms of CD13 were elucidated in liver cancer, thereby providing insights for the improved anticancer efficacy of CDDP. We present this article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-435/rc).
Methods
Reagents and antibodies
Ubenimex (purity ≥99.9%) was obtained from Main Luck (Shenzhen, China). CDDP was acquired from Jiangsu Hansoh (Shanghai, China). Cyclopamine (purity ≥98%) was purchased from Meilun Biotech (Dalian, China). 2-deoxy-D-glucose (2-DG, purity >99%) was obtained from Abmole (Houston, TX, USA). The following primary antibodies were used for western blot analysis: Mouse monoclonal antibodies against ANPEP/CD13 (1:5,000; cat. No. 66211-1-Ig; ProteinTech, Rosemont, IL, USA); rabbit polyclonal antibodies against PTCH1 (1:1,000; cat. No. E-AB-10571; Elabscience, Wuhan, China), GLI1 (1:1,000; cat. no: E-AB-19459; Elabscience), SMO (1:1,000; cat. No. DF5152; Affinity Biosciences), sonic hedgehog protein (SHH; 1:1,000; cat. No. DF7747; Affinity Biosciences), β-actin (1:1,000; cat. No. E-AB-20058; Elabscience), HK2 (1:1,000; cat. No. E-AB-14706; Elabscience), PKM2 (1:1,000; cat. No. AF5234; Affinity Biosciences), GLUT1 (1:1,000; cat. No. E-AB-31556; Elabscience). Horseradish peroxidase-conjugated anti-mouse (1:10,000; cat. No. E-AB-1001) and anti-rabbit (1:10,000; cat. No. E-AB-1003) secondary antibodies were obtained from Elabscience.
Cell culture
The liver cancer cell lines Huh7, HepG2, and PLC/PRF/5 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Huh7 and PLC/PRF/5 cells were grown in Dulbecco’s modified Eagle medium (DMEM, Procell Life Science & Technology, Wuhan, China) and HepG2 cells were cultured in modified Eagle’s medium (MEM, Procell Life Science & Technology). All cells were cultured in the aforementioned medium with 10% fetal bovine serum (FBS, Procell Life Science & Technology) in a humidified 5% CO2 incubator at 37 ℃. The pTZU-CD13-short hairpin RNA (shRNA) and p enhanced green fluorescent protein (EGFP)-N1-CD13 plasmids were purchased from GeneChem (Shanghai, China) and transfected into the cells by Lipofectamine™ 3000 (Thermo Fisher Scientific, Waltham, MA, USA).
Western blot analysis
RIPA buffer (Solarbio, Beijing, China) containing 1% protease and phosphatase inhibitors (Elabscience) was used to extract the total protein from the treated liver cancer cell lines. The concentration was measured by using the bicinchoninic acid method to estimate the loading amount. Following addition of the loading buffer to the protein, the latter was boiled and denatured. Electrophoresis was performed using a Mini-PROTEAN Tetra Cell system (Bio-Rad Laboratories Inc., Hercules, CA, USA). Following transfer of the desired protein to a polyvinylidene fluoride membrane (MilliporeSigma), blocking was performed using a 5% skimmed milk solution. The membranes were incubated with primary antibodies at 4 ℃ for 12–16 h. Subsequently, they were incubated with a homologous secondary antibody. ChemiDoc™ Imager (Fusion Solo; VilberLourmat, Collegien, France) was used to detect the band signals visualized by the ECL Chemiluminescence Kit (Epizyme, Shanghai, China).
Immunofluorescence staining
The liver cancer cells were seeded in a 12-well plate, treated, and fixed with 4% polyformaldehyde at room temperature for 15 min. The cells were washed with phosphate-buffered saline (PBS) three times and blocked with 5% goat serum blocking solution at room temperature for 45 min. Following blocking, the cells were exposed to the corresponding primary antibody (1:200) overnight at 4 ℃; they were washed with PBS three times and subsequently incubated with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (1:300) in the dark at room temperature for 1 h. Finally, an anti-fluorescent quencher containing 4',6-diamidino-2-phenylindole (DAPI) was added. Immunofluorescent images were captured by an inverted fluorescence microscope (NikonTi2-U; Nikon Corporation, Tokyo, Japan). ImageJ software (National Institutes of Health) was used to obtain the integrated optical density (OD) of the fluorescence intensity.
Enzyme-linked immunosorbent assay (ELISA)
The activity levels of the intracellular metabolic enzymes HK2 and PKM2 were detected with the corresponding ELISA kits (Elabscience) according to the manufacturer’s instructions. The OD was detected spectrophotometrically. The enzyme concentration levels were estimated by substituting the OD value into the standard curve formula.
Measurement of energy metabolic parameters
The intracellular levels of adenosine triphosphate (ATP) were detected using an ATP chemiluminescence assay kit (Elabscience) according to the manufacturer’s instructions. Luminescence was recorded using the luminometer. The levels of lactate in the culture supernatant were determined using the lactate colorimetric assay Kit (Elabscience) according to the protocol provided by the manufacturer.
Cell proliferation assay
The liver cancer cell lines were seeded in 96-well plates (5×103 cells/well) and treated in a humidified 5% CO2 incubator at 37 ℃ for 24, 48, and 72 h. At the indicated time points, the supernatant was replaced by 100 µL medium containing 10 µL Cell Counting Kit-8 (CCK-8) reagent (Yeasen, Shanghai, China) in each well. Subsequently, it was incubated for 1 h at 37 ℃. The OD450 value was detected using a microplate reader (SynergyMx; BioTek, Winooski, VT, USA).
Wound healing assay
The monolayer cells with 90% density were scratched vertically with a 10 µL pipette tip to retain the same scratch pattern in each well. Following washing with PBS for three times, the cells were incubated in a drug-containing medium with 2% FBS in a humidified 5% CO2 incubator at 37 ℃ for 48 h. The images were obtained by microscopy at 0, 24, and 48 h. The area of the cell migration was assessed using ImageJ software (National Institutes of Health).
Transwell assays
The transwell invasion experiments were performed as follows: The cells were incubated in starvation medium overnight. Matrigel matrix glue (Corning Inc., Corning, NY, USA) was placed at 4 ℃ for approximately 12 h to melt. A total of 50 µL matrigel diluted with serum-free DMEM (1:8) was added to the transwell chamber per well. Subsequently, it was placed in a 37 ℃ incubator for 1 h to solidify. A total of 1×105 cells were seeded with serum-free medium in the upper chamber. A total of 500 µL complete medium containing 10% serum was added to the 24-well plate. Following incubation for 48 h, the cells which were found on the membrane were carefully removed with cotton swabs, while the invasive cells under the membrane were fixed with 4% paraformaldehyde for 30 min at room temperature. Following washing with PBS, the cells were stained with 0.1% crystal violet for 20 min at room temperature. The migration experiment was carried out without matrigel. The stained cells on the bottom surface were observed using an inverted microscope (Nikon Ti2-U) and counted with ImageJ software (National Institutes of Health).
Cell apoptosis analysis by flow cytometry
The cells seeded in 6-well plates were treated with drugs for 48 h and digested using trypsin (Procell Life Science & Technology). The cells were centrifuged at 300 ×g for 5 min and washed twice with cold PBS. Following removal of the supernatant, the cells were resuspended in 1× binding buffer and stained with 5 µL FITC and 5 µL propidium iodide (PI) (BD Biosciences, San Jose, CA, USA) for 15 min at room temperature in the dark. Following filtering with 400 mesh sieves, the cells were detected by flow cytometry (CytoFLEX S; Beckman Coulter, Inc., Brea, CA, USA) and analyzed with FlowJo Software (BD Biosciences).
Statistical analysis
The data were presented as the mean ± standard deviation from triplicate experiments. GraphPad Prism software 7.0 (La Jolla, CA, USA, RRID:SCR_002798) was utilized for analysis. In order to analyze the differences between the two groups, the Student’s t-test was used, whereas one-way analysis of variance (ANOVA) was used for analysis of multiple groups and the comparison was made by Tukey’s method post hoc test. P<0.05 was considered statistically significant.
Results
Ubenimex inhibited aerobic glycolysis in liver cancer cell lines
Aerobic glycolysis is considered to possess a close relationship with the proliferation, invasion, and migration of liver cancer cell lines (9). The present study investigated whether ubenimex affects the glycolysis of liver cancer. The two key rate-limiting enzymes of glycolysis are HK2 and PKM2. ELISA experiments demonstrated that the enzyme activity levels of HK2 and PKM2 in liver cancer cell lines could be significantly inhibited by ubenimex, whereas the inhibition became more apparent with the increase of ubenimex dosage (Figure 1A,1B). Silencing of CD13 expression produced similar results (Figure 1C,1D). Simultaneously, western blot analyses illustrated that with the reduced expression of CD13, ubenimex could markedly decrease the expression levels of the key glycolytic proteins HK2, PKM2, and GLUT1 (Figure 1E-1G).
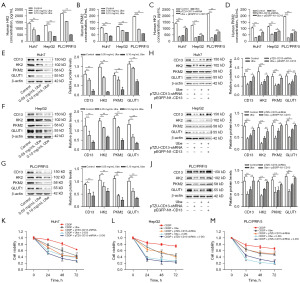
Consistent with these observations, the data demonstrated that CD13 silencing also suppressed the expression levels of the glycolysis-related proteins; ubenimex-mediated downregulation of HK2, PKM2, and GLUT1 protein levels was abolished by upregulation of CD13 expression (Figure 1H-1J). Moreover, the present study examined whether suppression of aerobic glycolysis could enhance the combined inhibition of ubenimex and CDDP on the proliferation of liver cancer cell lines. The glycolysis inhibitor 2-DG was applied to block aerobic glycolysis in liver cancer cell lines. As expected, the CCK-8 cell viability assay demonstrated that downregulation of CD13 expression or ubenimex treatment promoted the effects of CDDP; 2-DG further boosted the joint inhibitory effect caused by CDDP and ubenimex or by CD13 silencing on the proliferation of liver cancer cell lines (Figure 1K-1M). These data revealed that ubenimex significantly suppressed aerobic glycolysis in liver cancer cell lines.
Ubenimex blocked Hh signaling by targeting CD13 in liver cancer cell lines
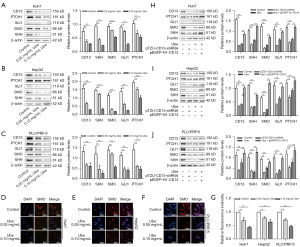
Hh signaling is crucial to the regulation of liver cancer; blocking Hh signaling can suppress cell proliferation (41). Since recent evidence suggests that glycolysis is activated by the Hh pathway in liver cancer cell lines, the role of CD13 in mediating Hh signaling was investigated in Huh7, HepG2, and PLC/PRF/5 cells. The effects of ubenimex on Hh signaling in Huh7, HepG2, and PLC/PRF/5 cells were initially assessed with western blot analysis. Consistent with the downregulation of the CD13 protein levels, western blot analysis confirmed that the levels of PTCH1, GLI1, SMO, and SHH proteins were significantly lower in ubenimex-treated liver cancer cell lines than in the control cells (Figure 2A-2C). These data illustrated that ubenimex dose-dependently inhibited Hh signaling in liver cancer cell lines. Moreover, immunofluorescence staining confirmed that ubenimex reduced the levels of SMO, which coincided with the previous conclusions (Figure 2D-2G). In addition, following ubenimex treatment or transfection of the cells with the pTZU-CD13-shRNA plasmid, the expression levels of PTCH1, GLI1, SMO, and SHH were downregulated, whereas the opposite effects were observed following overexpression of CD13 (Figure 2H-2J). Collectively, these findings indicated that ubenimex blocked Hh signaling by targeting CD13 in liver cancer cell lines.
Disruption of Hh signaling was essential for ubenimex inhibition of glycolysis liver cancer cell lines
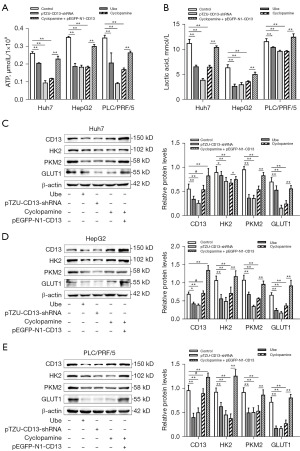
To determine whether ubenimex inhibits glycolysis by targeting the CD13/Hh pathway in liver cancer cell lines, the products of glycolysis including the intracellular levels of ATP and the levels of lactate in the culture supernatant were examined. Compared with the control group, ubenimex treatment and CD13 silencing reduced the production of ATP and lactate, which was similar to the inhibitory effects of the Hh inhibitor cyclopamine (Figure 3A,3B). Western blot analysis indicated that ubenimex, CD13 silencing, and cyclopamine suppressed the expression levels of HK2, PKM2, and GLUT1 (Figure 3C-3E). The expression levels of CD13 were downregulated with ubenimex treatment or following silencing of CD13 expression. However, the expression levels of the CD13 protein were not significantly affected by cyclopamine (Figure 3C-3E). These results demonstrated that inhibition of CD13 expression and suppression of the Hh pathway could decrease glycolysis of liver cancer cell lines; Hh signaling was the downstream pathway of CD13. Furthermore, upregulation of CD13 expression weakened the effects of ubenimex, CD13 silencing, and cyclopamine in reducing the expression of glycolysis-related proteins (Figure 3C-3E), which further illustrated that CD13 was an indispensable part of this mechanism. Taken together, these data revealed that ubenimex inhibited glycolysis by blocking CD13/Hh signaling in liver cancer cell lines.
The glycolysis of liver cancer cell lines was suppressed by ubenimex through relieving CD13/Hh signaling so as to facilitate apoptosis, inhibit invasion, migration, and proliferation and enhance the effect of CDDP
The biological function of the CD13/Hh pathway and glycolysis was studied in liver cancer cell lines. Wound healing assays indicated that the CDDP group had lower scratch healing rates than the control group; in addition, the mobility of CDDP combined with ubenimex, the downregulated levels of CD13, cyclopamine, or 2-DG were lower than those noted in the CDDP treatment alone group (Figure 4A-4E). In addition, the results of the transwell assay suggested that the number of the cells in the lower chamber following CDDP treatment was lower than that in the control group and higher than that in the other combined treatment groups (Figure 4F-4I). These results demonstrated that ubenimex enhanced the inhibitory effect of CDDP on the invasion and migration of liver cancer cell lines.
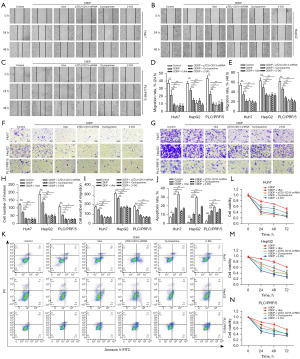
In addition to the aforementioned experiments, flow cytometry was performed to investigate the rate of apoptosis. The apoptosis assay results demonstrated that CDDP combined with ubenimex, CD13 silencing, cyclopamine, or 2-DG, enhanced apoptosis compared with the CDDP single treatment group (Figure 4J,4K). The data indicated that ubenimex enhanced the therapeutic effect of CDDP by promoting the induction of apoptosis in liver cancer cell lines.
Furthermore, based on the CCK-8 results, the cell viability of liver cancer cell lines was suppressed following treatment with CDDP. Cell viability was lower in the combined treatment groups (Figure 4L-4N). However, no significant difference was noted between the group treated with ubenimex and the group transfected with the pTZU-CD13-shRNA plasmid. Taken together, these findings indicated that ubenimex induced apoptosis and suppressed the biological function of liver cancer cell lines by inhibiting glycolysis via alleviating the CD13/Hh pathway, thus enhancing the effect of CDDP.
Discussion
As the sixth most common cancer globally, liver cancer ranks fifth in cancer-related morbidity and second in mortality in China. The prognosis of liver cancer is not optimistic due to its highly invasive metastasis and the high rate of postoperative recurrence. Due to the concealed onset of the disease and its rapid progression, patients are usually diagnosed at an advanced stage (2). Radiofrequency ablation (RFA) and transcatheter arterial chemoembolization (TACE) are Food and Drug Administration (FDA)-approved treatments for advanced HCC (3). Additional first line therapy for advanced HCC includes broad-spectrum tyrosine kinase inhibitors (TKIs), such as sorafenib and lenvatinib, as well as a combination of immunotherapy and anti-angiogenesis therapy, namely atezolizumab and bevacizumab (3,42,43). Research and development of targeted, safe and effective treatment mode can provide significant survival benefits for HCC patients. CD13 is overexpressed in various solid tumors and participates in complex and diverse processes, such as tumor cell invasion, differentiation, cell migration, and angiogenesis (44). Numerous studies have established that the CD13 inhibitor ubenimex can improve the efficacy of chemotherapy (35,37,39). Nonetheless, the ability of ubenimex to regulate glycolysis by targeting the CD13/Hh pathway so as to enhance the efficacy of CDDP has not been previously investigated. In the present study, ubenimex was shown to inhibit glycolysis.
Aerobic glycolysis ensures the necessary energy and materials required for the rapid tumor growth and metastasis of cancer cells, which offers advantageous environment for the development of cancer, including liver cancer (45,46). The changes in the expression levels of the glycolytic regulators (HK2, PKM2, and GLUT1) can greatly affect the progression of liver cancer and are associated with the pathological stages of liver cancer tumors (20,47,48). Liver cancer expresses considerably higher levels of HK2 than most adult tissues; silencing of its expression inhibits proliferation and increases sensitivity to cell death in human liver cancer cell lines (47,49). The expression levels of PKM2 are considered to be associated with the clinical and pathological features of liver cancer (50). Heat shock protein 90 (HSP90) promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance in HCC (51). As an important molecule which controls the rate of glucose transport, high levels of GLUT1 are associated with the proliferation and invasiveness of liver cancer (52). The present study indicated that the expression levels of the glycolytic key proteins correlated positively with the expression of CD13 in Huh7, HepG2, and PLC/PRF/5 cells. Following CD13 overexpression, the inhibition of glycolysis by ubenimex was reversed, further verifying that ubenimex regulated glycolysis by targeting CD13. In addition, the glycolysis inhibitor 2-DG further potentiated the inhibitory effects of ubenimex or the effects caused by synergism of downregulation of CD13 expression with CDDP treatment on the proliferation of liver cancer cell lines. In summary, the proliferation of liver cancer cell lines was inhibited by blockade of aerobic glycolysis, which was mediated by CD13. However, the exact mechanism requires further verification.
Hh signaling plays a central role in embryonic development, which is inactive in the adult liver, except for the involvement in post-injury reconstruction (53). The Hh signaling components are highly expressed in liver cancer tissues and cell lines (54-56). Previous study has shown that liver cancer cell lines secrete SHH via exosomes and promote tumorigenesis through the activated Hh pathway (22). Moreover, the knockdown of PHD finger protein 19 (PHF19) expression promotes the ubiquitination of GLI1 to activate the Hh signaling pathway and facilitate tumorigenesis of liver cancer (57). In addition, microRNA-338-3p inhibits invasion of liver cancer cell lines by regulating SMO (56). The high mRNA expression of PTCH1 and GLI1 in the SHH pathway is related to the high risk of recurrence of liver cancer following operation (58). In the current study, western blot analysis indicated that the expression levels of the Hh pathway ligands decreased with the decrease of CD13 expression, as demonstrated by the fluorescence intensity of SMO. Expectedly, these effects were attenuated by upregulation of CD13 expression, highlighting that the latter was required for the activation of the opposite pathway of Hh via ubenimex.
Accumulating evidence has shown that Hh signaling regulates glycolysis in several diseases, including liver cancer (9,33). In the present study, ubenimex was shown to regulate glycolysis and inhibit the Hh pathway. Therefore, it was speculated that ubenimex regulates the aerobic glycolysis of liver cancer cell lines via Hh signaling. The data of the present study revealed that CD13 silencing or ubenimex treatment significantly reduced not only the levels of ATP and lactate, but also the expression levels of HK2, PKM2, and GLUT1. Cyclopamine induced a similar effect; however, the effect was completely different following overexpression of CD13. Overall, ubenimex inhibited glycolysis mediated by CD13/Hh signaling in liver cancer cell lines.
According to the present study, the reduction in the expression levels of CD13 augmented the effect of CDDP in inhibiting the progression of liver cancer with regard to cell invasion, migration, proliferation, and apoptosis. Moreover, inhibition of Hh signaling or glycolysis can effectively assist CDDP in suppressing the progression of liver cancer, suggesting that ubenimex may play a synergistic role in inhibiting glycolysis by targeting the CD13/Hh pathway. However, this requires additional studies for confirmation.
Conclusions
To conclude, the aggregated data in this study revealed that ubenimex blocks Hh pathway and inhibits glycolysis in liver cancer cell lines. We also newly discovered that ubenimex facilitates apoptosis and suppresses the invasion, migration, and proliferation of liver cancer cell lines via regulating CD13/Hh signaling-mediated glycolysis to enhance the curative effect of CDDP. These findings outline the importance of CD13 in liver cancer. Due to the complexity of Hh pathway and metabolic mechanism, further research is needed to determine the detailed molecules involved.
Certainly, there are several limitations in our study. Experimental research in vivo is needed, as well as research on the specific molecular mechanisms between CD13 and Hh pathways in aerobic glycolysis of liver cancer cell lines. These restrictions will be further perfected in our future work.
Acknowledgments
We would like to thank Donna Pringle from the Oregon Health Sciences University, USA for reviewing the language and style of the manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-435/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-435/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-435/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-435/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Oura K, Morishita A, Tani J, et al. Tumor Immune Microenvironment and Immunosuppressive Therapy in Hepatocellular Carcinoma: A Review. Int J Mol Sci 2021;22:5801. [Crossref] [PubMed]
- Paskeh MDA, Ghadyani F, Hashemi M, et al. Biological impact and therapeutic perspective of targeting PI3K/Akt signaling in hepatocellular carcinoma: Promises and Challenges. Pharmacol Res 2023;187:106553. [Crossref] [PubMed]
- Chakraborty E, Sarkar D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers (Basel) 2022;14:2798. [Crossref] [PubMed]
- Sun Y, Wu L, Zhong Y, et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 2021;184:404-421.e16. [Crossref] [PubMed]
- Xu XF, Yang XK, Song Y, et al. Dysregulation of non-coding RNAs mediates cisplatin resistance in hepatocellular carcinoma and therapeutic strategies. Pharmacol Res 2022;176:105906. [Crossref] [PubMed]
- Moriguchi M, Umemura A, Itoh Y. Current status and future prospects of chemotherapy for advanced hepatocellular carcinoma. Clin J Gastroenterol 2016;9:184-90. [Crossref] [PubMed]
- Zou L, Sun P, Zhang L. miR-651-3p Enhances the Sensitivity of Hepatocellular Carcinoma to Cisplatin via Targeting ATG3-Mediated Cell Autophagy. J Oncol 2021;2021:5391977. [Crossref] [PubMed]
- WARBURG O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Feng J, Li J, Wu L, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res 2020;39:126. [Crossref] [PubMed]
- Wang Y, Chen X, Yao N, et al. MiR-448 suppresses cell proliferation and glycolysis of hepatocellular carcinoma through inhibiting IGF-1R expression. J Gastrointest Oncol 2022;13:355-67. [Crossref] [PubMed]
- Yu M, Chen S, Hong W, et al. Prognostic role of glycolysis for cancer outcome: evidence from 86 studies. J Cancer Res Clin Oncol 2019;145:967-99. [Crossref] [PubMed]
- Cui H, Gao Q, Zhang L, et al. Knockdown of FOXK1 suppresses liver cancer cell viability by inhibiting glycolysis. Life Sci 2018;213:66-73. [Crossref] [PubMed]
- Zhou W, Zhang S, Cai Z, et al. A glycolysis-related gene pairs signature predicts prognosis in patients with hepatocellular carcinoma. PeerJ 2020;8:e9944. [Crossref] [PubMed]
- Liu YC, Lin P, Zhao YJ, et al. Pan-cancer analysis of clinical significance and associated molecular features of glycolysis. Bioengineered 2021;12:4233-46. [Crossref] [PubMed]
- Deng T, Ye Q, Jin C, et al. Identification and validation of a glycolysis-associated multiomics prognostic model for hepatocellular carcinoma. Aging (Albany NY) 2021;13:7481-98. [Crossref] [PubMed]
- Wei Q, Ren Y, Zheng X, et al. Ginsenoside Rg3 and sorafenib combination therapy relieves the hepatocellular carcinoma progression through regulating the HK2-mediated glycolysis and PI3K/Akt signaling pathway. Bioengineered 2022;13:13919-28. [Crossref] [PubMed]
- Liang B, Jiang Y, Song S, et al. ASPP2 suppresses tumour growth and stemness characteristics in HCC by inhibiting Warburg effect via WNT/β-catenin/HK2 axis. J Cell Mol Med 2023;27:659-71. [Crossref] [PubMed]
- Zhu Q, Lei Z, Xu C, et al. LncRNA HClnc1 facilitates hepatocellular carcinoma progression by regulating PKM2 signaling and indicates poor survival outcome after hepatectomy. Cancer Med 2023;12:14526-44. [Crossref] [PubMed]
- Yao J, Tang S, Shi C, et al. Isoginkgetin, a potential CDK6 inhibitor, suppresses SLC2A1/GLUT1 enhancer activity to induce AMPK-ULK1-mediated cytotoxic autophagy in hepatocellular carcinoma. Autophagy 2023;19:1221-38. [Crossref] [PubMed]
- Zheng X, Gou Y, Jiang Z, et al. Icaritin-Induced FAM99A Affects GLUT1-Mediated Glycolysis via Regulating the JAK2/STAT3 Pathway in Hepatocellular Carcinoma. Front Oncol 2021;11:740557. [Crossref] [PubMed]
- Doheny D, Manore SG, Wong GL, et al. Hedgehog Signaling and Truncated GLI1 in Cancer. Cells 2020;9:2114. [Crossref] [PubMed]
- Li L, Zhao J, Zhang Q, et al. Cancer Cell-Derived Exosomes Promote HCC Tumorigenesis Through Hedgehog Pathway. Front Oncol 2021;11:756205. [Crossref] [PubMed]
- Kim J, Hyun J, Wang S, et al. Thymosin beta-4 regulates activation of hepatic stellate cells via hedgehog signaling. Sci Rep 2017;7:3815. [Crossref] [PubMed]
- Gu Y, Wang Y, He L, et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer 2021;20:132. [Crossref] [PubMed]
- Wu D, Yin YQ, Li Y, Zhang L, et al. CK2α causes stemness and chemotherapy resistance in liver cancer through the Hedgehog signaling pathway. Hepatobiliary Pancreat Dis Int 2023;22:383-91. [Crossref] [PubMed]
- Su GF, Huang ZX, Huang DL, et al. Cepharanthine hydrochloride inhibits the Wnt/β-catenin/Hedgehog signaling axis in liver cancer. Oncol Rep 2022;47:83. [Crossref] [PubMed]
- Che L, Yuan YH, Jia J, et al. Activation of sonic hedgehog signaling pathway is an independent potential prognosis predictor in human hepatocellular carcinoma patients. Chin J Cancer Res 2012;24:323-31. [Crossref] [PubMed]
- Chen Y, Choi SS, Michelotti GA, et al. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology 2012;143:1319-1329.e11. [Crossref] [PubMed]
- Ge X, Lyu P, Gu Y, et al. Sonic hedgehog stimulates glycolysis and proliferation of breast cancer cells: Modulation of PFKFB3 activation. Biochem Biophys Res Commun 2015;464:862-8. [Crossref] [PubMed]
- Lian N, Jiang Y, Zhang F, et al. Curcumin regulates cell fate and metabolism by inhibiting hedgehog signaling in hepatic stellate cells. Lab Invest 2015;95:790-803. [Crossref] [PubMed]
- Du K, Hyun J, Premont RT, et al. Hedgehog-YAP Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells. Gastroenterology 2018;154:1465-1479.e13. [Crossref] [PubMed]
- Hinshaw DC, Hanna A, Lama-Sherpa T, et al. Hedgehog Signaling Regulates Metabolism and Polarization of Mammary Tumor-Associated Macrophages. Cancer Res 2021;81:5425-37. [Crossref] [PubMed]
- Chen Y, Zhu W. Knockdown of the Sonic Hedgehog (SHH) Gene Inhibits Proliferation of Hep3B and SMMC-7721 Hepatocellular Carcinoma Cells via the PI3K/Akt/PCK1 Signaling Pathway. Med Sci Monit 2019;25:6023-33. [Crossref] [PubMed]
- Mina-Osorio P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med 2008;14:361-71. [Crossref] [PubMed]
- Guzman-Rojas L, Rangel R, Salameh A, et al. Cooperative effects of aminopeptidase N (CD13) expressed by nonmalignant and cancer cells within the tumor microenvironment. Proc Natl Acad Sci U S A 2012;109:1637-42. [Crossref] [PubMed]
- Guo Q, Sui ZG, Xu W, et al. Ubenimex suppresses Pim-3 kinase expression by targeting CD13 to reverse MDR in HCC cells. Oncotarget 2017;8:72652-65. [Crossref] [PubMed]
- Haraguchi N, Ishii H, Mimori K, et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest 2010;120:3326-39. [Crossref] [PubMed]
- Xiu T, Guo Q, Jing F, et al. CD13 downregulation mediated by ubenimex inhibits autophagy to overcome 5-FU resistance by disturbing the EMP3/FAK/NF-κB pathway in gastric cancer cells. Transl Cancer Res 2022;11:2487-500. [Crossref] [PubMed]
- Yamashita M, Wada H, Eguchi H, et al. A CD13 inhibitor, ubenimex, synergistically enhances the effects of anticancer drugs in hepatocellular carcinoma. Int J Oncol 2016;49:89-98. [Crossref] [PubMed]
- Guo Q, Jing FJ, Qu HJ, et al. Ubenimex Reverses MDR in Gastric Cancer Cells by Activating Caspase-3-Mediated Apoptosis and Suppressing the Expression of Membrane Transport Proteins. Biomed Res Int 2019;2019:4390839. [Crossref] [PubMed]
- Wang Y, Wang H, Yan Z, et al. The critical role of dysregulated Hh-FOXM1-TPX2 signaling in human hepatocellular carcinoma cell proliferation. Cell Commun Signal 2020;18:116. [Crossref] [PubMed]
- Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med 2020;382:1894-905. [Crossref] [PubMed]
- Yang C, Zhang H, Zhang L, et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2023;20:203-22. [Crossref] [PubMed]
- Pang L, Zhang N, Xia Y, et al. Serum APN/CD13 as a novel diagnostic and prognostic biomarker of pancreatic cancer. Oncotarget 2016;7:77854-64. [Crossref] [PubMed]
- Du D, Liu C, Qin M, et al. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm Sin B 2022;12:558-80. [Crossref] [PubMed]
- Zheng YL, Li L, Jia YX, et al. LINC01554-Mediated Glucose Metabolism Reprogramming Suppresses Tumorigenicity in Hepatocellular Carcinoma via Downregulating PKM2 Expression and Inhibiting Akt/mTOR Signaling Pathway. Theranostics 2019;9:796-810. [Crossref] [PubMed]
- DeWaal D, Nogueira V, Terry AR, et al. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat Commun 2018;9:446. [Crossref] [PubMed]
- Wong CC, Au SL, Tse AP, et al. Switching of pyruvate kinase isoform L to M2 promotes metabolic reprogramming in hepatocarcinogenesis. PLoS One 2014;9:e115036. [Crossref] [PubMed]
- Patra KC, Wang Q, Bhaskar PT, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013;24:213-28. [Crossref] [PubMed]
- Wong N, Ojo D, Yan J, et al. PKM2 contributes to cancer metabolism. Cancer Lett 2015;356:184-91. [Crossref] [PubMed]
- Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer 2017;16:178. [Crossref] [PubMed]
- Wang P, Sun J, Sun C, et al. BTF3 promotes proliferation and glycolysis in hepatocellular carcinoma by regulating GLUT1. Cancer Biol Ther 2023;24:2225884. [Crossref] [PubMed]
- Gao L, Zhang Z, Zhang P, et al. Role of canonical Hedgehog signaling pathway in liver. Int J Biol Sci 2018;14:1636-44. [Crossref] [PubMed]
- Huang S, He J, Zhang X, et al. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis 2006;27:1334-40. [Crossref] [PubMed]
- Kim Y, Yoon JW, Xiao X, et al. Selective down-regulation of glioma-associated oncogene 2 inhibits the proliferation of hepatocellular carcinoma cells. Cancer Res 2007;67:3583-93. [Crossref] [PubMed]
- Huang XH, Chen JS, Wang Q, et al. miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J Pathol 2011;225:463-72. [Crossref] [PubMed]
- Xiaoyun S, Yuyuan Z, Jie X, et al. PHF19 activates hedgehog signaling and promotes tumorigenesis in hepatocellular carcinoma. Exp Cell Res 2021;406:112690. [Crossref] [PubMed]
- Jeng KS, Sheen IS, Jeng WJ, et al. High expression of patched homolog-1 messenger RNA and glioma-associated oncogene-1 messenger RNA of sonic hedgehog signaling pathway indicates a risk of postresection recurrence of hepatocellular carcinoma. Ann Surg Oncol 2013;20:464-73. [Crossref] [PubMed]


