
Screening for mutations in lung cancer in France: purpose of precision medicine
In non-small cell lung cancer (NSCLC), the purpose of precision medicine is to use the latest genomic knowledge to adapt treatments to patients. It is essential that drugs are designed to hit a molecular abnormality, mutation or translocation, inducing NSCLC. Compared with other cancers, genetic alterations in NSCLC are notably high (1). In NSCLC, FDA and EMEA have already approved epidermal growth factor receptor (EGFR) inhibitors, gefitinib, erlotinib or afatinib, in the front line setting of EGFR mutated NSCLC and anaplastic lymphoma kinase (ALK) inhibitor, crizotinib, in ALK or ROS1 translocated NSCLC (2,3). Several novel cancer therapies targeting oncogenic mutations as BRAF or MET mutations may be approved in NSCLC in the next years.
The two major issues of precision medicine are the complex biology and the economic costs (4). Thus, targeted drugs need to be accompanied by valid diagnostic tests to identify patients who will benefit of these therapies. EGFR or ALK testing are cost saving as expensive drugs will be exclusively prescribed to patients who will gain benefit (5). However many health-care systems have no funding to pay for these tests.
Proceeding efforts are necessary in molecular dismantling of NSCLC to provide a tailored therapy to a maximum of patients. In France, prescription of EGFR or ALK targeting therapies are conditioned by molecular alterations and these testings are done routinely. In 2006, the French National Cancer Institute (INCa) has set up a national program to support molecular testing with the establishment of 28 regional molecular genetics centres. Screened molecular alterations were selected in 2009, including EGFR mutations, ALK gene rearrangements, but also emerging biomarkers such as KRAS, BRAF, HER2 or PI3KCA mutations. Furthermore, INCa developed a quality assurance program for molecular testing (ISO 15189).
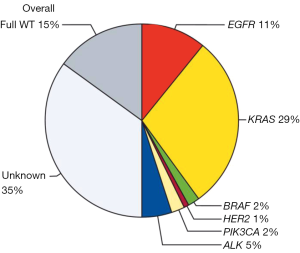
The BIOMARKER France study assessed the characteristics, molecular profiles and clinical outcomes of patients who were screened by this programme from 04/2012 to 04/2013. Data reported in Lancet on more than 17,000 patients show the presence of at least one genetic alteration in about 50% of analysed samples (6). Thus, EGFR mutations were detected in 11% of samples, HER2 mutations un 1%, KRAS mutations in 29%, BRAF mutations in 2% and PI3K mutations in 2% of patients; ALK rearrangements were detected in 5% of the analysed samples (Figure 1). The presence of a genetic alteration affected first line treatment for 51% of patients with a significant improvement in the proportion of patients achieving an overall response in the first line or second line treatment and an improved overall survival [16.5 months (15.0–18.3 months) versus without a genetic alteration 11.8 months (10.1–13.5 months); P<0.0001]. However improved prognosis in NSCLC harbouring EGFR mutations or ALK rearrangements compared to wild-type NSCLC is reported. Thus whether this effect on overall survival is related to specific medications such as EGFR and ALK inhibitors (predictive) or to the prognosis of NSCLC is hypothetical. This systematic biomarker analysis was greeted as a major innovation by ASCO in 2013 (7).
This French project, as well as other initiatives as the German Network Genomic Medecine (NGM), the national wide Japanese Lung Cancer Screening Network (LC-SCRUM) and the American Lung Cancer Mutational Consortium (LCMC), participate to a better understanding of NSCLC.
In the BIOMARKER France study, no improvement in the inclusion rate of clinical trials was noticed; thus only 3% of patients with a molecular alteration were included in a clinical trial. Molecular alterations were selected in 2009 and emerging biomarkers such as KRAS, HER2, BRAF and PI3KCA mutations were routinely analyzed, also for these molecular abnormalities, no targeted therapies were available. Data on targeting HER2 or BRAF mutations are now robust (8,9). It is not certain that KRAS or PI3KCA are effective targets for tailored therapy and whether these mutations should be routinely detected is speculative. ROS1 testing and MET amplification/mutations are now part of the routine molecular testing on the molecular platforms. Since 2014, INCa supports ACSé program to assess the effectiveness of crizotinib in MET amplified/mutated or ROS rearranged and vemurafenib in BRAF mutated NSCLC (9-11). Further large scale molecular screening studies should collaborate with pharmaceutical companies to target emerging biomarkers. Thus, the Japanese LC-SCRUM study includes a genomic analysis by next generation sequencing multiplexing diagnostics and a collaboration with 13 pharmaceutical companies to deliver drugs on the basis of the patients genomic alteration (12).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shao-Hua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.05.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- . Getting personal. Nature 2011;473:253-4. [Crossref] [PubMed]
- Borget I, Cadranel J, Pignon JP, et al. Cost-effectiveness of three strategies for second-line erlotinib initiation in nonsmall-cell lung cancer: the ERMETIC study part 3. Eur Respir J 2012;39:172-9. [Crossref] [PubMed]
- Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. [Crossref] [PubMed]
- Patel JD, Krilov L, Adams S, et al. Clinical Cancer Advances 2013: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J Clin Oncol 2014;32:129-60. [Crossref] [PubMed]
- Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997-2003. [Crossref] [PubMed]
- Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAFV600E-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. [Crossref] [PubMed]
- Moro-Sibilot D, Faivre L, Zalcman G, et al. Crizotinib in patients with advanced ROS1-rearranged non-small cell lung cancer (NSCLC). Preliminary results of the ACSé phase II trial. J Clin Oncol;2015;33:abstr 8065.
- Vassal G, Ledeley MC, Tournigand C, et al. Activity of crizotinib in relapsed MET amplified malignancies: Results of the French AcSé Program. J Clin Oncol 2015;33:abstr 2595.
- Matsumoto S, Tsuchihara K, Yoh K, et al. A new nationwide genomic screening system in Japan for the development of targeted therapies against advanced non-small lung cancers with rare driver mutations. J Clin Oncol 2014;32:abstr 11007.


