
Low OGDHL expression affects the prognosis and immune infiltration of kidney renal clear cell carcinoma
Highlight box
Key findings
• Oxoglutarate dehydrogenase-like (OGDHL) can be used as an independent risk factor to predict kidney renal clear cell carcinoma (KIRC). Low OGDHL is associated with poor prognosis and immunity in KIRC.
What is known and what is new?
• Low OGDHL expression is associated with poor prognosis in cancers, including liver, pancreatic and cervical cancer, etc.
• A prognostic model of OGDHL in KIRC was constructed to explore the mechanisms associated with immune infiltration and carcinogenesis.
What is the implication, and what should change now?
• OGDHL shows promise as a new target for KIRC immunotherapy.
Introduction
Renal cell carcinoma (RCC) is a prevalent form of cancer worldwide, with approximately 430,000 new cases annually (1,2). The predominant histological subtype of RCC, kidney renal clear cell carcinoma (KIRC), accounts for approximately 70% of all cases (3). Unfortunately, KIRC lacks reliable early diagnostic indicators, and imaging techniques such as contrast-enhanced tomography or magnetic resonance imaging are required for detection (4). Furthermore, numerous newly diagnosed patients with KIRC are already in advanced stages of the disease, often presenting with distant metastases and displaying limited response to conventional chemotherapy and radiotherapy. Despite significant advances in targeted therapy such as tyrosine kinase inhibitors and mammalian target of rapamycin (mTOR) inhibitors, as well as immunotherapy with programmed cell death protein 1/programmed cell death ligand 1 and cytotoxic T-lymphocyte-associated antigen 4 inhibitors, patients with advanced and metastatic KIRC continue to face an unsatisfactory prognosis (5,6). Therefore, it is important to identify meaningful prognostic biomarkers and elucidate the molecular mechanisms underlying KIRC to identify new therapeutic targets.
The development of cancer cells is intricately linked to energy metabolism, with glutamine serving as the main carbon source after glucose during energy production and anabolism (7). Glutamine promotes mitochondrial metabolism to sustain the proliferation of cancer cells and inhibiting this metabolic process has been shown to effectively induce anticancer effects (8,9). Oxoglutarate dehydrogenase-like (OGDHL), a subunit of the oxoglutarate dehydrogenase complex (OGDHC), is a crucial component involved in glucose and glutamic acid degradation in the tricarboxylic acid cycle, and is associated with energy metabolism in cancer (10,11). Reduction in OGDHL expression or inhibition of OGDHC activity would lead to a decrease in glutamine metabolism and an increase in glutamine dependence in cancer cells, which is related to poor liver cancer prognosis (12). Liu et al. showed that OGDHL regulated the microRNA-214/TWIST1 pathway and inhibited the growth and metastasis of pancreatic cancer (13). Sen et al. found that OGDHL knockdown would activate the protein kinase B (AKT) signaling pathway to promote cervical carcinogenesis (14). Guo et al. reported that breast cancer morbidity was associated with rare coding variants of OGDHL (15). Mao et al. also showed that low OGDHL expression was associated with poor prognosis in thyroid cancer (16).
However, an in-depth exploration of the biological significance and prognostic implication of OGDHL in KIRC is yet to be conducted. In the present study, we compared the differential expression of OGDHL in normal and tumor tissues by analyzing multiple databases, evaluating its diagnostic value, analyzing the correlation between clinical features and survival prognosis, determining independent prognostic factors, and studying the role and biological function of OGDHL in tumor microenvironment (TME). Our ultimate objective was to elucidate the correlation between OGDHL expression prognosis, and immune infiltration in KIRC, with the aim of identifying novel biomarkers that can serve as targets for future therapeutic interventions. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-961/rc).
Methods
Data collection
The Cancer Genome Atlas (TCGA) gene expression profiles and clinical characteristics of 535 KIRC tumor and 72 normal samples were obtained from the University of California Santa Cruz Xena database (http://xena.ucsc.edu/). The information of 522 patients with KIRC was selected for further analysis by matching the gene expression data and clinical information. In addition, we incorporated data from the Gene Expression Omnibus database, specifically the GSE53757 dataset (17). This dataset encompasses tumor specimens and their matched normal tissues from a cohort of 72 patients diagnosed with KIRC. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene Expression Profiling Interactive Analysis (GEPIA2) and Tumor Immune Estimation Resource (TIMER)
GEPIA2 is an online gene expression analysis tool of considerable versatility, enabling insights into diverse gene expression levels across both tumor and normal tissue contexts (18). TIMER, is an extensively acknowledged biological information analysis tool renowned for its substantial functionality, garnered substantial recognition within scholarly circles (19). GEPIA2 was used exclusively to analyze the differential expression patterns of OGDHL in both normal and KIRC contexts, whereas TIMER primarily focused on comprehending the broader implications across various cancer types. Both tools were used to corroborate their impact on the survival prognosis of patients with KIRC. Within the foundational framework of these methodologies, adjustments were made solely pertaining to gene nomenclature and the specific cancer types of interest, while retaining the default system parameters for both tools.
Human Protein Atlas (HPA) and The University of Alabama at Birmingham Cancer (UALCAN)
The HPA serves as a discriminating tool to discern the variance in protein expression between normal and pathological tissues using immunohistochemistry (20). UALCAN provided a quantitative approach for corroborating protein expression profiles (21). We validated the expression levels of the OGDHL protein using two distinct websites.
Construction of prognostic model
This study used the log2[fragments per kilobase million (FPKM) +1] data format to investigate the prognostic significance of OGDHL expression in tumor samples. Receiver operating characteristic (ROC) curve analysis was performed to assess the diagnostic value of OGDHL for KIRC. Additionally, the correlation between OGDHL expression and clinical features was analyzed, and both univariate and multivariate Cox regression analyses were performed to determine the independent risk factors associated with the disease. The criteria for all high- and low-expression groups were defined based on the median expression level of OGDHL in KIRC samples.
Evaluation of immune infiltration
The CIBERSORT algorithm was used to assess the relative abundance of 22 tumor-infiltrating immune cells (TIICs) across 522 tumor samples. Specifically, the proportion of TIICs in groups with high and low OGDHL expression was compared. Furthermore, the proportions of TIICs in normal and tumor tissues were compared. The CIBERSORT algorithm uses the LM22 gene set (https://cibersortx.stanford.edu/) to generate a matrix of annotated gene sets that defines 22 distinct immune cell subtypes.
Gene set enrichment analysis (GSEA)
In our analysis, we included a list of genes that exhibited an expression difference of more than two-fold between normal and KIRC samples (P<0.05) (table available at https://cdn.amegroups.cn/static/public/tcr-23-961-1.xlsx). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to investigate the potential mechanisms or pathways associated with OGDHL and KIRC, including biological processes, cell components, molecular function, and ten OGDHL-associated signaling pathways. In addition, the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) was used to stratify the samples into two groups based on the median expression of OGDHL, and GSEA was used to explore the activation or inhibition of specific mechanisms or pathways.
Statistical analysis
Data processing and statistical analysis were performed using R software v4.2.2 (http://www.r-project.org/) and the corresponding R package, GraphPad Prism 8.0.2. The present study employed the statistical t-test to assess potential discrepancies in OGDHL gene expression across distinct groups. The P value of the Kaplan-Meier survival curve was calculated using the log-rank test. Chi-squared test was used to evaluate the correlation between OGDHL gene expression and clinical features. Univariate and multivariate Cox regression analyses were used to identify independent prognostic factors. Statistical significance was set as P<0.05.
Results
OGDHL expression was significantly down-regulated in KIRC tissues
We first performed transcript differential expression analysis using DESeq2 and determined that compared with normal tissues, OGDHL was down-regulated by 2-fold in KIRC tissues (Padj<0.05; Figure 1A). To clarify the specific reason for the difference, we extracted and analyzed the expression levels of the OGDHL gene in tumor and normal samples. As shown in the results, OGDHL expression was significantly down-regulated in tumor samples (P<0.001; Figure 1B). In addition, to verify the reliability of the results, we used two online databases, GEPIA2 and TIMER, from which the same results were obtained, with statistical significance (P<0.05; Figure 1C,1D). We used the GSE53757 dataset for investigation (17). The outcomes unequivocally demonstrated the robust validation of OGDHL within this dataset (P<0.001; Figure 1E). To investigate the disparities in the protein-level expression of OGDHL, immunohistochemical observations were conducted using the HPA website. The staining intensity of KIRC specimens was notably diminished compared with that of normal tissues (Figure 1F,1G). Similarly, the UALCAN website effectively corroborated the downregulation of OGDHL expression in cancerous tissues (P<0.001; Figure 1H).

The prognostic significance and diagnostic potential of OGDHL in KIRC
To investigate the effect of OGDHL on the survival of patients with KIRC, we conducted a thorough analysis of TCGA-KIRC cohort. We eliminated four duplicate cases and meticulously performed Kaplan-Meier survival analysis on 522 cancer cases that possessed OGDHL expression data alongside clinical information. The results indicated that patients with low OGDHL expression had poor overall survival (OS) (P<0.001; Figure 2A). To ensure that the results were reliable, we again obtained the same results through analysis of the GEPIA2 and TIMER online databases (P<0.001; Figure 2B,2C). These results strongly suggest that OGDHL is a good predictor of survival in KIRC. We plotted an ROC curve to assess the value of OGDHL in the diagnosis of KIRC. The analysis showed that OGDHL was highly sensitive in the diagnosis of KIRC and had reliable diagnostic value [area under the curve (AUC) =0.917; 95% confidence interval (CI): 0.885–0.949; Figure 2D].
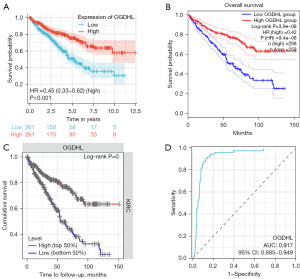
Correlation between OGDHL expression and clinical features
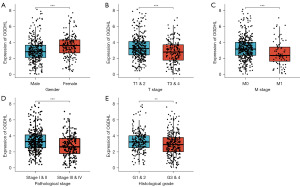
We used the median OGDHL expression as the critical point, and 522 patients with KIRC with clinical information matching were divided into two groups (high and low OGDHL expression) to determine their relationship with clinicopathological features. Low OGDHL expression was significantly associated with the T stage (P=0.018), M stage (P<0.001), pathological stage (P=0.001), histological grade (P=0.018), and sex (P<0.001) (Table 1). These factors may play an important role in the prognosis of KIRC; therefore, we conducted an additional stratified analysis. The results showed that there were significant differences in OGDHL expression levels between different sexes (P<0.001; Figure 3A), which was consistent with the higher incidence of KIRC in males than in females (1). The low expression of OGDHL was significantly correlated with the progression of T and M stages (T1 and 2 vs. T3 and 4, P<0.001, Figure 3B; M0 vs. M1, Figure 3C), and the expression of OGDHL decreased significantly with the progression of tumor pathology, and the expression in stage III and IV was significantly lower than that in stage I and II (P<0.001; Figure 3D). Moreover, a similar trend was observed for tumor histological grade (G1 and 2 vs. G3 and 4, P<0.01; Figure 3E). These results revealed a significant correlation between low OGDHL expression and poor prognosis in patients with KIRC.
Table 1
| Characteristic | Total | High (n=261), n (%) | Low (n=261), n (%) | P |
|---|---|---|---|---|
| Age | 522 | 0.661 | ||
| <60 years | 124 (47.5) | 118 (45.2) | ||
| ≥60 years | 137 (52.5) | 143 (54.8) | ||
| Gender | 522 | <0.001 | ||
| Female | 122 (46.7) | 59 (22.6) | ||
| Male | 139 (53.3) | 202 (77.4) | ||
| T stage | 522 | 0.018 | ||
| T1 & 2 | 180 (69.0) | 153 (58.6) | ||
| T3 & 4 | 81 (31.0) | 108 (41.4) | ||
| N stage | 254 | 0.256 | ||
| N0 | 132 (95.7) | 106 (91.4) | ||
| N1 | 6 (4.3) | 10 (8.6) | ||
| M stage | 492 | <0.001 | ||
| M0 | 218 (90.1) | 196 (78.4) | ||
| M1 | 24 (9.9) | 54 (21.6) | ||
| Pathological stage | 519 | 0.001 | ||
| Stage I & II | 176 (68.0) | 139 (53.5) | ||
| Stage III & IV | 83 (32.0) | 121 (46.5) | ||
| Histological grade | 514 | 0.018 | ||
| G1 & 2 | 131 (51.4) | 105 (40.5) | ||
| G3 & 4 | 124 (48.6) | 154 (59.5) |
OGDHL, oxoglutarate dehydrogenase-like; KIRC, kidney renal clear cell carcinoma.
Prognostic value of OGDHL in KIRC
Previously, we found that patients with different OGDHL expression levels have significantly different OS according to the Kaplan-Meier survival analysis. Further investigations are warranted to gain a deeper understanding of the factors associated with the prognosis of KIRC. Univariate and multivariate Cox regression analyses were performed on important clinical features (sex, age, pathological stage, and histological grade) and OGDHL expression levels (Table 2). Patients <60 years of age had a significantly lower risk of death than patients ≥60 years of age [hazard ratio (HR) =0.696, 95% CI: 0.504–0.961, P=0.027]. The progression of pathological stage, histological grade and low OGDHL expression were independently associated with OS, and the risk of death was significantly increased (HR =2.791, 95% CI: 1.987–3.922, P<0.001; HR =1.948, 95% CI: 1.351–2.809, P<0.001; HR =2.127, 95% CI: 1.511–2.994, P<0.001). Sex did not significantly affect the prognosis, but the difference was no statistical significance. These results indicate that age, pathological stage, histological grade and OGDHL expression are independent prognostic factors for KIRC.
Table 2
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |||
| Age | 522 | |||||
| ≥60 years | 280 | Reference | ||||
| <60 years | 242 | 0.563 (0.411–0.772) | <0.001 | 0.696 (0.504–0.961) | 0.027 | |
| Gender | 522 | |||||
| Male | 341 | Reference | ||||
| Female | 181 | 1.054 (0.772–1.440) | 0.740 | 1.375 (0.976–1.938) | 0.069 | |
| Pathological stage | 519 | |||||
| Stage I & II | 315 | Reference | ||||
| Stage III & IV | 204 | 3.927 (2.847–5.417) | <0.001 | 2.791 (1.987–3.922) | <0.001 | |
| Histological stage | 514 | |||||
| G1 & 2 | 236 | Reference | ||||
| G3 & 4 | 278 | 2.679 (1.895–3.787) | <0.001 | 1.948 (1.351–2.809) | <0.001 | |
| OGDHL | 522 | |||||
| High | 261 | Reference | ||||
| Low | 261 | 2.198 (1.605–3.009) | <0.001 | 2.127 (1.511–2.994) | <0.001 | |
KIRC, kidney renal clear cell carcinoma; CI, confidence interval; OGDHL, oxoglutarate dehydrogenase-like.
Subsequently, we stratified these prognostic factors. The results showed that when age ≥60 years (P=0.001), age <60 years (P<0.001), patients were male (P<0.001), G3 and 4 (P<0.001), the OS of patients with low OGDHL expression was significantly lower than high OGDHL expression, and no further statistical significant differences were observed (Figure 4A-4D).

Correlation analysis between OGDHL expression and tumor infiltrating immune cells
The degree of immune cell infiltration in tumor tissues is closely related to patient prognosis. Díaz-Montero et al. have shown that KIRC possesses a high degree of immunogenicity, which leads to immune dysfunction by inducing immunosuppressive cell infiltration (22). However, the relationship between OGDHL expression and tumor infiltrating immune cells in KIRC is not yet fully understood. We first analyzed the proportion of 22 TIICs from TCGA-KIRC samples using the CIBERSORT algorithm. The results showed a difference in the proportion of TIICs in each sample (Figure 5A). Subsequently, differences in the proportion of TIICs types between the two groups (high and low OGDHL expression) were further analyzed. The results showed significant differences in the proportions of the eight TIICs types (Figure 5B). Plasma cells (P<0.05), T cells gamma delta (P<0.05), CD4+ memory resting T cells (P<0.05), natural killer (NK) cells activated (P<0.05), CD4+ activated cells (P<0.001), resting mast cells (P<0.001), and eosinophils (P<0.05). Therefore, the identification of dissimilar proportions of TIICs alone does not provide a comprehensive explanation for the prognostic implications in KIRC, and thus necessitates further stratified investigations.

Prognostic value of TIICs in KIRC
In the above results (Figure 5B), we identified several TIICs with statistically significant differences in proportion and reviewed the relevant literature using PubMed. Finally, we selected two types of TIICs, resting mast cells and T cells CD4 memory activated, to focus on the prognostic value of KIRC (23,24). The results indicated that lower levels of resting mast cells were significantly associated with worse OS (P=0.001, Figure 6A), histological grade (P<0.001, Figure 6B) and pathological stage (P<0.001, Figure 6C) deterioration, whereas T cells CD4 memory activated showed the opposite (P=0.021, Figure 6D; P<0.01, Figure 6E; P<0.05, Figure 6F).

OGDHL functional enrichment analysis
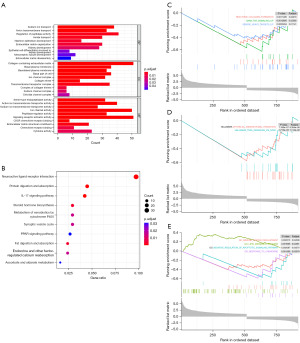
To further explore the function of OGDHL, GO and KEGG functional enrichment analysis were performed using the RStudio. GO functional enrichment analysis showed that OGDHL may be significantly related to peptidase activity, transmembrane transport, signal transduction, extracellular matrix, kidney formation, and epithelial cell differentiation involved in kidney development (Figure 7A). KEGG analysis indicated that substance metabolism, signal transduction, interleukin (IL)-17 and peroxisome proliferator-activated receptor (PPAR) signaling pathways were significantly enriched (Figure 7B). Next, we performed GSEA between the high and low OGDHL expression groups using multiple gene sets in the MSigDB database to better define the pathways of activation or repression. The results showed that multiple signaling pathways were significantly activated in the low OGDHL expression group, including epithelial-mesenchymal transition (EMT), tumor necrosis factor alpha (TNFα)/nuclear factor kappa B (NF-κB) signaling pathway, negative regulation of apoptotic signaling pathway, collagen formation, chemokine response and HDAC3 targets up. However, immune system development was inhibited (Figure 7C-7E).
Discussion
KIRC is a malignant neoplasm that, poses a significant threat to the human health. Despite remarkable progress in the study of KIRC treatment, patient prognosis remains suboptimal (6). Therefore, there is an imperative need to identify novel biomarkers that can serve as therapeutic targets in KIRC. However, the role of OGDHL in KIRC remained unclear.
In our study, we used TCGA, GEPIA2 and TIMER databases to verify that OGDHL was significantly down-regulated in KIRC. Consistent with this observation, other findings were obtained from the GSE53757 dataset (17). Subsequent validation using HPA and UALCAN revealed a marked reduction in the protein expression of OGDHL in neoplastic tissue. Finally, through survival analysis, diminished expression of OGDHL was substantiated as a harbinger of adverse OS outcomes. These findings provide a preliminary glimpse into the potential of OGDHL to function as a putative tumor suppressor gene, exerting its influence on the pathogenesis of KIRC. Other studies, have confirmed the inhibitory effect of OGDHL in other cancers, including liver, pancreatic, cervical, breast, and thyroid cancer (12-16). To assist in the diagnosis, the diagnostic value of OGDHL was assessed using ROC curves, and the results were promising, with OGDHL performing sensitively for the diagnosis of KIRC (AUC =0.917). Next, we analyzed the correlation between OGDHL and the clinical characteristics of patients with KIRC. The results showed that the expression level of OGDHL was negatively correlated with pathological features that could reflect the deterioration process (e.g., pathological stage and histological grade). More importantly, we found from univariate and multifactorial Cox analyses that OGDHL expression levels were an independent prognostic predictor of OS, which was confirmed in a stratified analysis based on age, sex and histological grade. However, there was no significant difference in survival between patients with stage I and II and stage III and IV, which could possibly be attributed to the relatively lower degree of malignancy of tumors in the stage I and II groups, whereas in the stage III and IV groups, tumors exhibited a high degree of invasiveness. Despite these unsatisfactory results, low expression of OGDHL generally indicates, a poor prognosis for patients with KIRC and is a prognostic biomarker worthy of recognition.
Malignant tumor development is intricately intertwined with metabolic reprogramming, wherein glutamine is a primary carbon source for cancer cell energy metabolism (7). Presently, RCC is widely acknowledged as a metabolic disorder, with alterations in energy metabolism pathways implicated in adverse prognostic outcomes, including increased cancer cells uptake of glutamine (25). Furthermore, studies have reported that glutathione is the chief product of the reprogrammed metabolism of glutamine (26,27). Lucarelli et al. elucidated that within KIRC tissue, nearly all amino acids, except save for glutamine and glutamic acid, exhibit substantial reduction, potentially linked to an increase in glutathione. Glutathione confers an advantage to cancer cells in the oxidative stress-laden TME, thus underscoring the potential of targeting intermediates in glutamine metabolism, such as α-ketoglutarate, as a latent therapeutic strategy (28). Previous studies corroborated the pivotal role of OGDHL in glutamine degradation. Diminished OGDHL expression orchestrates a heightened α-ketoglutarate/citrate ratio, thereby mediating the reductive carboxylation of glutamine, reinforcing the cancer cells antioxidative machinery, and in turn, fostering proliferation and survival (12). Collectively, these findings suggest the prospective utility of OGDHL as a prognostic biomarker and a potential therapeutic target in KIRC.
Cancer development is closely related to the TME. TIICs play a crucial role in KIRC (22). We used CIBERSORT to analyze the immune infiltration results of KIRC and found that the expression of OGDHL was positively correlated with the proportion of resting mast cells, whereas the opposite was true for the activation of T cells CD4+ memory was the opposite. Dudeck et al. reported that mast cells are an important part of the immune sentinel, that can enhance T cell-mediated immune response and exert anti-tumor effects (29). It has been reported that mast cells can also inhibit cancer by mediating the expression of anti-tumor heparin, histamine, IL-6 and TNFα (30). Our study verified this finding, which provides a direction for future treatment strategies for KIRC (Figure 6A-6C). The anti-tumor effect of CD4+ memory T cells have been widely reported (31). However, our results were not consistent with this finding, although, the opposite was true (Figure 6D-6F). Feuerer et al. also reported that the proportion of CD4+ memory T cells in the bone marrow of patients with breast cancer was notably higher than that in healthy donors, and the increase in memory T cells was related to the size of the primary tumor (32). This may be interpreted as an attempt by the immune system to attack the growing number of cancer cells in patients. However, there is still a lack of sufficient evidence, and it is worth considering the causes and underlying mechanisms. Tumor angiogenesis exhibits a conspicuous nexus with the intricate milieu of the TME (33). Angiogenesis is the principal hallmark of KIRC and plays a central role in various stages of progression. Anti-angiogenic therapy has evolved into an indispensable approach for addressing KIRC (34). Reportedly, approximately 92% of patients with KIRC exhibit somatic mutations in the Von Hippel-Lindau (VHL) gene. This process activates of numerous biological pathways, including pro-angiogenic factors (VEGF and PDGF) and genes involved in metabolism, thereby influencing the TME. Existing research has substantiated a notable elevation in the KIRC utilization of glutamine. Moreover, glutamine deprivation therapy has the potential to emerge as a novel treatment for individuals with VHL-deficient RCC (28,35). The orchestration of metabolic regulatory strategies has also been reported to engender transformative alterations in the TME. Inquisitors have marshaled glutaminase inhibitors that obstruct the synthesis of metabolites and abrogate cellular proliferation (36). As a pivotal rate-limiting enzyme in glutamine metabolism, OGDHL may influence the TME patterns by inhibiting angiogenesis or modulating glutamine metabolism, thereby affecting KIRC progression. Therefore, further investigations are necessary.
However, the potential function of OGDHL in KIRC has not been reported. We used the TCGA-KIRC database for GO and KEGG analyses of OGDHL-co-expressed genes and elucidated several mechanisms related to OGDHL, including extracellular matrix formation, signal transduction, kidney formation and epithelial cell differentiation, which are involved in kidney development (Figure 7A,7B). As a matter of common knowledge, KIRC is a malignant tumor originating from renal tubular epithelium (3). Notably, GO analysis showed that OGDHL was associated with epithelial cell differentiation, which is involved in kidney development (Figure 7A), indicating that OGDHL is closely related to the occurrence of KIRC.
We also performed GSEA to confirm whether signaling pathways were activated or inhibited by OGDHL in KIRC. The results showed that several important signaling pathways were significantly activated in the OGDHL low expression group, such as EMT, TNFα/NF-κB signaling pathway, negative regulation of apoptotic signaling pathway, collagen formation, HDAC3 targets up, and chemokine response signaling pathway activation (Figure 7C-7E). In the field of pathology, it has been observed that KIRC cells have an origin in renal tubular epithelial cells, and the occurrence, invasion, metastasis, and therapeutic resistance of malignancies are associated with the phenomenon of EMT (3,37). Chemokines significantly influence the TME, play a dual role in regulating the development and progression of cancer by mediating the transportation of immune cells to the TME and affecting the proliferation, metastasis, and invasion of tumor cells by targeting non-immune cells within the TME, including tumor cells and vascular endothelial cells (38). In tumors, the extracellular matrix (ECM) specific to the tumor is known to replace the normal ECM and exhibit greater collagen density and rigidity. Collagen, a major component of the ECM, has been shown to not only promote cancer cells growth and migration but also affect immune cell function within the TME (39). The regulation of apoptosis is an important way for the human body to eliminate cancer cells. However, tumor cells resist apoptotic signals or evolve various mechanisms to evade apoptosis, eventually leading to treatment resistance and recurrence (40). Our results showed that low OGDHL expression activated the negative regulation of apoptotic signals, which was associated with a poor prognosis in KIRC. In addition, we found that low OGDHL expression up-regulated of HDAC3 expression. Histone deacetylases (HDACs) play a crucial role in regulating cellular functions such as cell survival and proliferation through their ability to deacetylate histones. Aberrant expression of HDACs has been observed in various types of cancers. Wei et al. reported that HDAC3 was highly expressed in cancer cells and significantly promoted breast cancer (41). Zhang et al. showed that targeting HDAC3 could inhibit glutamine uptake by cancer cells, thereby promoting tumor regression (42). Another study demonstrated the feasibility of achieving the same goal by targeting the glutamine transporter protein SLC1A5 (43). As mentioned earlier, cancer cells growth is highly dependent on glutamine, and the downregulation of OGDHL reduces glutamine metabolism by inhibiting OGDHC activity while promoting increased dependence on glutamine by cancer cells, possibly through the involvement of HDAC3 or the glutamine transporter protein SLC1A5 (7,12,43). However, this hypothesis remains speculative and requires substantiation through experimental investigations. Our results also found that the activation of TNFα/NF-κB signaling pathway was associated with low OGDHL expression. TNFα/NF-κB as a classical pathway, its role in cancer has been widely and deeply studied, and it is not discussed here (44,45).
Although the present study highlights the potential impact of OGDHL on the prognosis of KIRC, several limitations must be acknowledged. Firstly, the findings are primarily derived from TCGA database; therefore, external validation is required. Second, the mechanism underlying the higher proportion of CD4+ memory T cells in KIRC tumor tissues than in normal tissues remains unclear and requires further investigation. Additionally, the specific effects of OGDHL on HDAC3 and the glutamine transporter SLC1A5 should be explored. Importantly, the influence of OGDHL on tumor angiogenesis or the modulation of glutamine metabolism, thereby inducing alterations in specific mechanisms of the TME, is a research avenue requiring in-depth exploration. Further fundamental research is required to elucidate the precise role of OGDHL in renal clear cell carcinoma.
Conclusions
Our study shows that OGDHL expression is significant in the diagnosis of KIRC and that reduced OGDHL expression is related to poor prognosis and immune infiltration in patients with KIRC. OGDHL is a novel biomarker of KIRC and is expected to become a therapeutic target in the future.
Acknowledgments
We thank the Xena platform, GSE53757, GEPIA2, TIMER, HPA, UALCAN, CIBERSORT and MSigDB databases for data support. We are also grateful to the Editage platform for checking and modifying the language.
Funding: This study was supported by Natural Science Foundation of Guangdong Province-General Project (No. 2020A1515010015), Social Development Technology Plan Project of Meizhou City (No.2020B090), and Cultivation Program of Meizhou People’s Hospital (No. PY-C 2020B090).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-961/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-961/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-961/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014;349:g4797. [Crossref] [PubMed]
- Gray RE, Harris GT. Renal Cell Carcinoma: Diagnosis and Management. Am Fam Physician 2019;99:179-84. [PubMed]
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur Urol 2022;82:399-410. [Crossref] [PubMed]
- Makhov P, Joshi S, Ghatalia P, et al. Resistance to Systemic Therapies in Clear Cell Renal Cell Carcinoma: Mechanisms and Management Strategies. Mol Cancer Ther 2018;17:1355-64. [Crossref] [PubMed]
- Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009;458:762-5. [Crossref] [PubMed]
- Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013;496:101-5. [Crossref] [PubMed]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 2010;35:427-33. [Crossref] [PubMed]
- Bunik V, Kaehne T, Degtyarev D, et al. Novel isoenzyme of 2-oxoglutarate dehydrogenase is identified in brain, but not in heart. FEBS J 2008;275:4990-5006. [Crossref] [PubMed]
- Bunik VI, Degtyarev D. Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins 2008;71:874-90. [Crossref] [PubMed]
- Dai W, Xu L, Yu X, et al. OGDHL silencing promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J Hepatol 2020;72:909-23. [Crossref] [PubMed]
- Liu Y, Meng F, Wang J, et al. A Novel Oxoglutarate Dehydrogenase-Like Mediated miR-214/TWIST1 Negative Feedback Loop Inhibits Pancreatic Cancer Growth and Metastasis. Clin Cancer Res 2019;25:5407-21. [Crossref] [PubMed]
- Sen T, Sen N, Noordhuis MG, et al. OGDHL is a modifier of AKT-dependent signaling and NF-κB function. PLoS One 2012;7:e48770. [Crossref] [PubMed]
- Guo X, Long J, Chen Z, et al. Discovery of rare coding variants in OGDHL and BRCA2 in relation to breast cancer risk in Chinese women. Int J Cancer 2020;146:2175-81. [Crossref] [PubMed]
- Mao M, Huang RZ, Zheng J, et al. OGDHL closely associates with tumor microenvironment and can serve as a prognostic biomarker for papillary thyroid cancer. Cancer Med 2021;10:728-36. [Crossref] [PubMed]
- von Roemeling CA, Radisky DC, Marlow LA, et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res 2014;74:4796-810. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci 2018;27:233-44. [Crossref] [PubMed]
- Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022;25:18-27. [Crossref] [PubMed]
- Díaz-Montero CM, Rini BI, Finke JH. The immunology of renal cell carcinoma. Nat Rev Nephrol 2020;16:721-35. [Crossref] [PubMed]
- Su Y, Zhang T, Lu J, et al. Identification and Validation of the Prognostic Panel in Clear Cell Renal Cell Carcinoma Based on Resting Mast Cells for Prediction of Distant Metastasis and Immunotherapy Response. Cells 2023;12:180. [Crossref] [PubMed]
- Chen L, Yin L, Qi Z, et al. Gene expression-based immune infiltration analyses of renal cancer and their associations with survival outcome. BMC Cancer 2021;21:595. [Crossref] [PubMed]
- Lucarelli G, Loizzo D, Franzin R, et al. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev Mol Diagn 2019;19:397-407. [Crossref] [PubMed]
- Lucarelli G, Rutigliano M, Sallustio F, et al. Integrated multi-omics characterization reveals a distinctive metabolic signature and the role of NDUFA4L2 in promoting angiogenesis, chemoresistance, and mitochondrial dysfunction in clear cell renal cell carcinoma. Aging (Albany NY) 2018;10:3957-85. [Crossref] [PubMed]
- Abu Aboud O, Habib SL, Trott J, et al. Glutamine Addiction in Kidney Cancer Suppresses Oxidative Stress and Can Be Exploited for Real-Time Imaging. Cancer Res 2017;77:6746-58. [Crossref] [PubMed]
- Lucarelli G, Rutigliano M, Loizzo D, et al. MUC1 Tissue Expression and Its Soluble Form CA15-3 Identify a Clear Cell Renal Cell Carcinoma with Distinct Metabolic Profile and Poor Clinical Outcome. Int J Mol Sci 2022;23:13968. [Crossref] [PubMed]
- Dudeck A, Köberle M, Goldmann O, et al. Mast cells as protectors of health. J Allergy Clin Immunol 2019;144:S4-S18. [Crossref] [PubMed]
- Kaesler S, Wölbing F, Kempf WE, et al. Targeting tumor-resident mast cells for effective anti-melanoma immune responses. JCI Insight 2019;4:e125057. [Crossref] [PubMed]
- Liu Q, Sun Z, Chen L. Memory T cells: strategies for optimizing tumor immunotherapy. Protein Cell 2020;11:549-64. [Crossref] [PubMed]
- Feuerer M, Rocha M, Bai L, et al. Enrichment of memory T cells and other profound immunological changes in the bone marrow from untreated breast cancer patients. Int J Cancer 2001;92:96-105. [Crossref] [PubMed]
- Xu Z, Xu J, Sun S, et al. Mecheliolide elicits ROS-mediated ERS driven immunogenic cell death in hepatocellular carcinoma. Redox Biol 2022;54:102351. [Crossref] [PubMed]
- Lucarelli G, Netti GS, Rutigliano M, et al. MUC1 Expression Affects the Immunoflogosis in Renal Cell Carcinoma Microenvironment through Complement System Activation and Immune Infiltrate Modulation. Int J Mol Sci 2023;24:4814. [Crossref] [PubMed]
- Lasorsa F, Rutigliano M, Milella M, et al. Cellular and Molecular Players in the Tumor Microenvironment of Renal Cell Carcinoma. J Clin Med 2023;12:3888. [Crossref] [PubMed]
- Vuong L, Kotecha RR, Voss MH, et al. Tumor Microenvironment Dynamics in Clear-Cell Renal Cell Carcinoma. Cancer Discov 2019;9:1349-57. [Crossref] [PubMed]
- Pastushenko I, Brisebarre A, Sifrim A, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556:463-8. [Crossref] [PubMed]
- Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17:559-72. [Crossref] [PubMed]
- Rømer AMA, Thorseth ML, Madsen DH. Immune Modulatory Properties of Collagen in Cancer. Front Immunol 2021;12:791453. [Crossref] [PubMed]
- Tong X, Tang R, Xiao M, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol 2022;15:174. [Crossref] [PubMed]
- Wei H, Ma W, Lu X, et al. KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression. Cancer Commun (Lond) 2021;41:904-20. [Crossref] [PubMed]
- Zhang HL, Chen P, Yan HX, et al. Targeting mTORC2/HDAC3 Inhibits Stemness of Liver Cancer Cells Against Glutamine Starvation. Adv Sci (Weinh) 2022;9:e2103887. [Crossref] [PubMed]
- Schulte ML, Fu A, Zhao P, et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med 2018;24:194-202. [Crossref] [PubMed]
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer 2009;9:361-71. [Crossref] [PubMed]
- Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer 2013;12:86. [Crossref] [PubMed]


