
Role for ubiquitin-specific protease 7 (USP7) in the treatment and the immune response to hepatocellular carcinoma: potential mechanisms
Introduction
Liver cancer is a common human tumor of the digestive system, cancer of the liver a serious global health issue (1,2). Liver cancer is the sixth most frequently diagnosed form of cancer worldwide and the fourth-leading cause of cancer deaths globally (3). It is well known that primary hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC) and mixed type (HCC-ICC) are the three most histologically heterogeneous malignant diseases. Among them, HCC is the most common primary hepatic carcinoma, representing approximately 90% of all liver cancers (4,5). Currently, hepatectomy with liver transplantation are the main treatment modalities. However, only 20–30% of patients have the chance for surgical resection, because a large proportion of patients have intermediate or advanced stage at diagnosis or have Child-Pugh grade C, and the postoperative recurrence rate is high, with short overall survival and poor quality of life (6). China is a high-risk area for HCC, with a higher incidence of HCC than other countries and a relatively poor 5-year survival rate (7). In order to reduce the burden of HCC, effective measures for diagnosis, treatment, and prognosis of HCC are needed (8). Biomarkers that can aid in diagnosis, guide treatment, and assess prognosis are under investigation, with a continued need for adjuvant therapy that prevents recurrence (9). Numerous studies have demonstrated immune cell infiltration of tumors to have the capacity to kill cancer cells (10-13). However, malignant cancer cells can evade the immune system (14). Analysis of immune cells infiltrating liver cell carcinoma may offer novel therapeutic approaches for the treatment of HCC (15).
Deubiquitinating enzymes (DUBs) regulate the stability, interaction, and localization of most cellular proteins by removal of ubiquitin modifications. Ubiquitin-specific protease 7 (USP7) is a member of the deubiquitinylase family of enzymes that removes ubiquitin from proteins and prevents substrate degradation. USP7 is involved in cancer development and is a promising target for cancer therapy (16,17). HCC tissues expressing high levels of USP7 undergo tumor growth, invasion, and are associated with poor HCC overall survival rates (18). These observations suggest that USP7 may be a possible diagnostic and therapeutic target for the management of malignant neoplasms of the liver. This study will evaluate this possibility (19,20). We present this article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-153/rc).
Methods
Data collection
USP7 gene expression data and clinically relevant information for HCC patients were downloaded in April 2022 from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/), the Gene Expression Omnibus (GEO) database (GSE10143; https://www.ncbi.nlm.nih.gov/geo/) and the International Cancer Genome Consortium database (ICGC; https://icgc.org/). HCC and paraneoplastic tissues from primary liver cancer inpatients at Affiliated Hospital of Nantong University from June to November 2022 were collected for external validation by immunohistochemistry and reverse transcription-polymerase chain reaction (RT-PCR) experiments. Inclusion criteria: (I) age >18 years; (II) meeting the diagnostic criteria of liver cancer, confirmed by liver function two-to-one, ultrasound and imaging; (III) complete clinical data; (IV) informed consent of patients. Exclusion criteria: (I) mental abnormalities, coagulation abnormalities or autoimmune system diseases; (II) combination of other malignant tumours; (III) cognitive impairment, recent radiotherapy and bioimmunotherapy; (IV) patients with incomplete clinical information. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Affiliated Hospital of Nantong University (No. 2023-L038). Informed consent was taken from all the patients.
Data processing and statistical analysis
Our data downloaded from TCGA, GEO, and ICGC were analysed using R software (https://www.r-project.org/). Immunohistochemistry and RT-PCR experimental methods of liver cancer patient tissue was used for external validation, which was analysed using SPSS AU. The UALCAN website (http://ualcan.path.uab.edu/analysis-prot.html) was also used to assess the correlation of USP7 expression levels with USP7 expression levels in HCC and tumour-adjacent tissues.
Functional pathway enrichment of USP7 in HCC by GSEA, GO, and KEGG analysis
Relevant pathways were explored by performing USP7 gene enrichment analysis using gene set enrichment analysis (GSEA) and the gene set “c2.cp.kegg.v7.4.symbols.gmt” of the Kyoto Encyclopedia of Genes and Genomes (KEGG). Pathways with a |normalized enrichment score (NES)| >1.5 and a nominal P<0.05 were selected for analysis. Gene Ontology (GO) analysis was used (c5.go.v7.4.symbols.gmt) to identify possible USP7 function and processes in HCC. Results suggest that USP7-related pathways may be related to the mechanism of HCC development and progression.
Correlation analysis of USP7 and immune function
We used single-gene pan-cancer analysis tools to evaluate the relationships among USP7 expression and differentially methylated probes-based (DMPss), epigenetically regulated DNA methylation-based (EREG-METHss), DNA methylation-based (DNAss), enhancer elements/DNAmethylation-based (ENHss), epigenetically regulated RNA expression-based (EREG.EXPss), RNA expression-based (RNAss), homologous recombination deficiency (HRD), and loss of heterozygosity (LOH) using the Sangerbox website (http://www.sangerbox.com/tool) by the Pearson method. We explored the relationship between USP7 expression levels and immune cells with the R language, while exploring the relationship between USP7 expression levels and immune checkpoint molecules. The relationship between USP7 expression and the level of immune cell infiltration was assessed using the Timer website. The effect of USP7 expression and immune cell infiltration on patient survival was investigated.
Relationship of drug sensitivity with USP7
We analyzed relationships of USP7 gene expression with the half inhibitory drug concentration (IC50) by use of the Genomics of Drug Sensitivity in Cancer (GDSC) database. The effect of USP7 expression was evaluated with regard to drug treatment regimen, USP7 inhibitor treatment, and in combination with other drugs. We assessed the relationship between sensitivity to nine common drugs and USP7 expression levels. At the same time, we mapped the mechanism of action of these nine drugs, combined with the effect of USP7 on drug sensitivity, and suggested possible targets of action of USP7.
Results
USP7 expression and diagnostic value for HCC
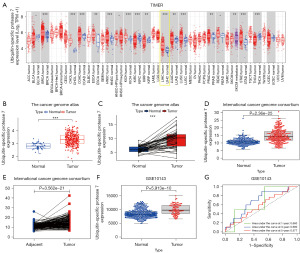
USP7 gene expression levels in TCGA tumor tissue and adjacent non-tumor tissue were evaluated and found to be markedly different (Figure 1A). The bar chart demonstrates USP7 expression to be observably higher in HCC tumors than in normal tissues (P<0.001; Figure 1B). The two box plots demonstrate USP7 mRNA expression to be generally elevated in tumor tissue compared to normal tissue (P<0.001; Figure 1C). USP7 mRNA expression was validated with ICGC and GSE10143 datasets and results were found to be similar to TCGA, with USP7 mRNA expression up-regulated in HCC tumor tissues (all P<0.001; Figure 1D-1F). We assessed the HCC diagnostic value of USP7 by receiver operating characteristic (ROC) analysis. Area under the curve (AUC) values for 1, 3, and 5 years were 0.660, 0.680, and 0.577, respectively. These results indicate that USP7 has a moderate diagnostic value (Figure 1G). In summary, USP7 is highly expressed in HCC and has a moderate diagnostic value for HCC, suggesting that the gene likely contributes to HCC development.
External verification of HCC associated USP7 protein and relationship of USP7 expression with clinico-pathology
We validated HCC USP7 protein with the UALCAN website by Clinical Proteomic Tumor Analysis Consortium (CPTAC) analysis and found total USP7 protein to be greater in primary HCC than in adjacent non-tumor hepatic tissue (P<0.001; Figure 2A). Figure 2B-2D identifies USP7 protein levels based on gender, age, and subtype 2. Immunohistochemistry and RT-PCR were performed to verify these results in cancer and adjacent tissues of HCC patients (Figure 2E-2G). The results demonstrate USP7 protein levels to be consistent with mRNA expression, and to be greater in tumor than in surrounding non-tumor liver tissue. We used R language to assess the relationships among USP7 mRNA expression and clinico-pathological features of HCC patients. USP7 mRNA expression was related to N and T stages (all P<0.05; Figure 2H-2J). Poor-expression of USP7 was related to stages 1 and T1. As such, HCC USP7 expression was greater during the later stages of the disease.

Construction of protein interaction networks
To explore the relationships among USP7 protein and the other proteins, we used R language to derive protein-protein interactive (PPI) networks for USP7 protein (Figure 3A-3F) as a means which assess the role of USP7 in the development of HCC. Eleven most important proteins were identified including: CREBBP, GLYR1, GSPT1, C16orf72, CEP20, EEF2K, ETFP, ATP5MPL, MRPL54, HSCB, and ITIH4. Among them, CREBBP is a transcriptional activator that plays a significant role in physiological processes such as embryonic development and growth control (21). EEF2K proteins regulate tumorigenesis and are up-regulated in different cancers, associated with patient prognosis, and are potential new tumor targets (22). The impact of these protein interactions with USP7 deserves further investigation.

USP7-related signaling pathways identified by GSEA
We divided USP7 mRNA expression into increased and decreased expression groups by GSEA to identify USP7-associated signaling pathways. We identified seven signaling pathways differentially enriched for high USP-7 expression; JAK-STAT, MAPK, P53, VEGF, WNT, CHEMOKINE, and CANCER signaling pathways (Figure 4A-4H). The JAK-STAT signaling pathway is involved in numerous biological processes. It is associated with many diseases, and is involved in malignant tumors and autoimmune disease (23,24). Many JAK inhibitors have been used in clinical practice with good curative results. The P53 signaling pathway is involved in tumor inhibition, immunological responses, and modulation of ferroptosis (25). Aberrant regulation of the WNT signaling pathway is linked to cancer biology. Inhibition of WNT signaling can hinder cancer cell growth and even kill cancer cells (26). VEGF plays a key role in tumor angiogenesis, influencing tumor cell growth (27). The MAPK/ERK signaling pathway regulates a variety of cellular processes, and it is frequently activated in human HCC cases (28,29). These pathways are associated with various physiological processes such as cell growth, differentiation, and apoptosis. Identification of signaling pathways associated with the USP7 gene in HCC provides new insight into the pathogenesis of HCC.

GO and KEGG analysis
We explored the potential role of USP7 in HCC by performing GO and KEGG analysis of USP7 co-expressed genes. Results found USP7 co-expressed genes to be primarily involved in ion channel regulation and cell signaling transduction pathways (Figure 5A-5E). Signal transduction and ion transport pathways of cells are related to; cell proliferation, differentiation, growth, senescence, death, metabolism, nerve conduction, immunity, and other basic life activities. The ID for GO: 0007188 access is the adenylate cyclase (AC) regulated G protein coupled receptor signaling pathway. AC is an integral membrane protein that converts ATP into cAMP as a form of cell signaling. AC is also an effector of the G protein coupling system. G protein-coupled receptors (GPCRs) participate in a number of physiological processes such as regulation of the autonomic nervous system, cell density, maintenance of the steady state, and are therapeutic target families (30). Indeed, the development of multiple cancers has been related to a functional lack of GPCRs (31). Therefore, GPCRs are expected to become tumor therapeutic targets and to occupy an increasingly important position in cancer research.

USP7 gene expression and immunity
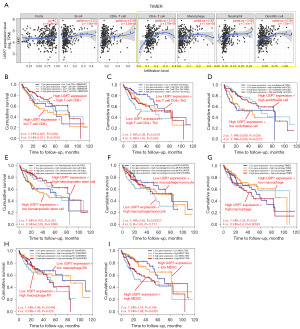
Results have shown USP7 to be significantly associated with neuropilin-1 (NRP1) in HCC (Figure 6A,6B). Immune checkpoint inhibitors are powerful cancer immune therapeutic (32). NRP1 is known to limit CD8+T cell-mediated antitumor immune responses during anti-PD1 immunotherapy (33). Further, USP7 inhibitors have been shown to target NRP1 immune checkpoint molecules, killing tumor cells. We assessed the relationship between USP7 and the HCC tumor microenvironment using the cell infiltration algorithms; EPIC, QUANTISEQ, MCPcounter, CIBERSORT, Xcell, and TIMER. We found CD4+ T cells, natural-killer (NK) cells, regulatory T (Tregs) cells, T-helper 1 (Th1) cells, macrophages, macrophage M2 cells, neutrophils, basophils, mesenchymal stem cells, smooth muscle cells, and dendritic cells to be significantly related to USP7 expression (Figure 6C-6H). Further, different levels of immune cell infiltration combined with USP7 expression levels were associated with HCC patient survival (Figure 7).

We used Sangerbox to calculate six tumor dryness indices, mRNA expression, and methylation signature to assess the relationships among dryness and gene expression by tumor cells (Figure 8). A higher score indicates more stem cells and a lower score indicates tumor differentiation. We found USP7 gene expression to be positively related to hepatoma; DMPss, DNAss, ENHss, and EREG-methss and negatively related to Ereg, EXPss, and RNAss. As such, USP7 expression was positively related to DNA methylation based stemness scores, and negatively related to RNA based stemness scores.

By analysis of the relationships among genomic heterogeneity and gene expression, we found USP7 gene expression to be positively related to HRD and LOH in HCC. HRD status is a key indicator for various tumor treatment options and prognosis, as well as highly associated with platinum-based chemotherapeutic drugs and poly ADP-ribose polymerase (PARP) inhibitor sensitivity. LOH is a chromosomal event capable of causing the loss of entire genes and their nearby chromosomal regions. LOH has been implicated in malignant tumorigenesis.
Therapeutic value of the USP7 gene for HCC
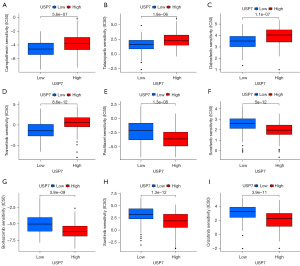
We investigated the relationships among USP7 gene expression and the IC50 using the GDSC database (Figure 9). It is well known that IC50 is an indicator of the antitumor activity of a particular drug (34), indicating approximately one half of a drug amount necessary to inhibit for example; enzymes, cell receptors, or microorganisms. IC50 can be used to assess the ability of a drug to induce apoptosis, with the greater the drug’s ability to induce apoptosis, the lower the value. The figure below shows that the greater the IC50, the higher the expression resistance. HCC patients with high USP7 gene expression were resistant to; camptothecin, talazoparib, dabrafenib, and trametinib. Paclitaxel, sorafenib, bortezomib, sunitinib, and crizotinib patients were more sensitive to greater USP7 gene expression. We assessed the mechanism of action of nine drugs (Figure 10) and found that high levels of USP7 expression attenuated drug effects for; BRAF, MEK, TOPOI, and PARP. USP7 inhibitors are known to induce tumor cell apoptosis. Therefore, tumor cell apoptosis resulting from USP7 inhibitor administration may be related to; B-Raf proto-oncogene serine/threonine protein kinase (BRAF), mitogen-activated extracellular signal-regulated kinase (MEK), DNA topoisomerase I (TOPOI), and PARP inhibitors. These observations require further experimental verification. In summary: (I) USP7 may act directly or indirectly on these four drug targets. (II) USP7 inhibitors can enhance the antitumor effects of these four drugs, suggesting that inhibition of these four targets in combination with USP7 inhibitors may have more powerful antitumor effects than monotherapy.

Discussion
Liver cancer is a common tumor of the human digestive system and a global health challenge (35). HCC has a poor prognosis, is highly invasive, and a leading cause of cancer-related deaths worldwide (36). HCC is currently the fourth most common cause of cancer death (37). Abnormal gene expression is associated with the development of HCC, affecting diagnosis, treatment, and prognosis (38). With the increasing burden of liver cancer disease, new biomarkers are urgently needed to provide for earlier diagnosis, improved treatment, and better prognosis. The treatments for early-stage liver cancer are local ablation, surgical resection, and liver transplantation (39). Immune checkpoint inhibitors are currently certified as an effective treatment option for patients with advanced cancer (40). Liver cancer monitoring increases the possibility of early detection and treatment. HCC accounts for 90% of liver cancer and is the most common primary hepatic carcinoma (41,42). USP7 expression in liver cell carcinoma is substantially higher than in adjacent normal tissue, so exploration of its use as a biomarker for HCC diagnosis, therapy, and prognosis is appropriate (43-45).
It is well known that ubiquitination is involved in the regulation of protein quantity and quality, regulating a variety of physiological activities, such as cell proliferation, differentiation, apoptosis, the immune response, and inflammation. Further, ubiquitination and deubiquitination enzymes are involved in the development of malignant tumors. USP7, also known as herpes virus-related ubiquitin-specific protease (HAUSP), has been found to be associated with many diseases. Therefore, we explored the expression level of USP7 in HCC, evaluating its therapeutic and immune value, as well as its mechanism of action. Our findings demonstrate USP7 expression to be significantly higher in primary carcinoma of the liver than in non-tumor tissue. USP7 expression was validated with the ICGC dataset, the GSE10143 dataset, and by immunohistochemistry. USP7 expression was significantly related to the N and T stages of disease. GSEA identified signaling pathways related to USP7 including: JAK-STAT, MAPK, P53, VEGF, and WNT. Furthermore, we found USP7 expression to be primarily associated with immune cells, immune checkpoint molecules, genomic heterogeneity, and tumor stemness. We found HCC patient expression levels of USP7 to predict survival as well as infiltration of immune cell populations.
Previously, the USP7 gene was shown to be closely associated with tumor-related proteins and proteins of the immune system including: phosphatase and tensin homologue (PTEN), forkhead box (FOXO4), NOTCH1, and DNMT1. PTEN is a tumor suppressor associated with tumor aggressiveness, wherein deubiquitination of PTEN by USP7 plays a pivotal role in PTEN localization and function. There is a direct relationship between USP7 level and nuclear rejection of PTEN, such that USP7 affects tumor aggressiveness and is associated with advanced cancer. USP7 is closely associated with ovarian cancer, chronic lymphocytic leukemia, myeloma, breast cancer, and prostate cancer. USP7 has been shown to induce prostate cancer cell migration and invasion by stabilizing the Enhancer of Zeste homolog 2 (EZH2) gene. Inhibition of USP7 reduces oncogene function, augments tumor suppression, and sensitizes tumors to DNA damaging agents. Abnormal activation or over expression of USP7 facilitates tumor development, suggesting the importance of timely therapeutic intervention.
In agreement with a previous publication, our results demonstrate USP7 expression to be greater in HCC than in normal tissue, and that USP7 expression modulates immune cells. By GSEA, we identified seven signaling pathways associated with USP7 including: JAK-STAT, MAPK, P53, VEGF, WNT, CHEMOKINE and CANCER pathways. It has been reported that JAK-STAT, MAPK, and P53 signaling pathways are closely associated with many important biological processes including cellular activity as well as immune regulation (28,29). VEGF receptors can promote the formation of tumor neovascularization, enabling tumors to obtain sufficient nutrients for tumor growth. The WNT signaling pathway is a complex network of proteins and plays a significant role in tumor progression and development (27,46).
Overall, USP7 expression was found to be significantly associated with immune cells and immune checkpoint molecules. Immune cells play an important role in tumor protection and their infiltration can predict tumor immunotherapy outcomes. Moreover, immune checkpoint suppression has become a key focus for cancer therapy, aimed at assisting the immune system in the identification and attack of cancer cells. Compared with conventional therapies, immune checkpoint suppression therapy can provide rapid and durable treatment effects for those patients with cancer, especially those with advanced metastatic cancer.
High levels of USP7 expression were found in tumor tissues by TCGA database analysis. This result was validated with the ICGC dataset, GSE10143 dataset, and immunohistochemistry. USP7 expression is not only relevant to immunity but is also associated with partial drug sensitivity, which may assist drug treatment selection. The mechanism of USP7 inhibitor-induced apoptosis of tumor cells may be related to targets, such as BRAF, MEK, TOPOI and PARP.
There are limitations to this study. First, patient treatment information was not considered. Second, we confirmed the importance of only a few cancer-related signaling pathways by GSEA; JAK-STAT, MAPK, P53, VEGF, and WNT pathways. These pathways were enriched for patients with high levels of USP7. Indeed, the results only indicate a USP7 connection with these pathways. Further investigations are necessary to confirm these results. Third, the relationship of high USP7 gene expression and therapeutic drug sensitivity was demonstrated for four drug targets; BRAF, MEK, TOPOI and PARP. The role of each of these and USP7 in HCC requires further investigation. Future investigations should explore the relationship of USP7 mRNA with tissues and immune-related genes.
Conclusions
In conclusion, this study identifies an oncogenic role for USP7 in HCC. GSEA identified USP7-related pathways including: JAK-STAT, MAPK, P53, VEGF, and WNT. USP7 was found to be significantly related to immunity and to partial drug sensitivity. The mechanism of tumor cell apoptosis induced by USP7 inhibitors was found to be related to; BRAF, MEK, TOPOI and PARP. Finally, USP7 inhibitors were shown to enhance antitumor effects against these four targets, although further experimental verification is necessary.
Acknowledgments
Funding: This research was funded by the China Postdoctoral Science Foundation (No. 2020M670041ZX) and the Nantong Science and Technology Project (No. HS2020001).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-153/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-153/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-153/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-153/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Affiliated Hospital of Nantong University (No. 2023-L038). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. [Crossref] [PubMed]
- Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res 2021;149:1-61. [Crossref] [PubMed]
- Zheng R, Qu C, Zhang S, et al. Liver cancer incidence and mortality in China: Temporal trends and projections to 2030. Chin J Cancer Res 2018;30:571-9. [Crossref] [PubMed]
- Sia D, Villanueva A, Friedman SL, et al. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017;152:745-61. [Crossref] [PubMed]
- Liu YC, Yeh CT, Lin KH. Cancer Stem Cell Functions in Hepatocellular Carcinoma and Comprehensive Therapeutic Strategies. Cells 2020;9:1331. [Crossref] [PubMed]
- McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021;73:4-13. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Kim E, Viatour P. Hepatocellular carcinoma: old friends and new tricks. Exp Mol Med 2020;52:1898-907. [Crossref] [PubMed]
- Nguyen PHD, Wasser M, Tan CT, et al. Trajectory of immune evasion and cancer progression in hepatocellular carcinoma. Nat Commun 2022;13:1441. [Crossref] [PubMed]
- Mao X, Xu J, Wang W, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 2021;20:131. [Crossref] [PubMed]
- Fan C, Zhang S, Gong Z, et al. Emerging role of metabolic reprogramming in tumor immune evasion and immunotherapy. Sci China Life Sci 2021;64:534-47. [Crossref] [PubMed]
- Kohli K, Pillarisetty VG, Kim TS. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther 2022;29:10-21. [Crossref] [PubMed]
- Anwanwan D, Singh SK, Singh S, et al. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer 2020;1873:188314. [Crossref] [PubMed]
- Zhang Q, Lou Y, Bai XL, et al. Immunometabolism: A novel perspective of liver cancer microenvironment and its influence on tumor progression. World J Gastroenterol 2018;24:3500-12. [Crossref] [PubMed]
- Nininahazwe L, Liu B, He C, et al. The emerging nature of Ubiquitin-specific protease 7 (USP7): a new target in cancer therapy. Drug Discov Today 2021;26:490-502. [Crossref] [PubMed]
- Li P, Liu HM. Recent advances in the development of ubiquitin-specific-processing protease 7 (USP7) inhibitors. Eur J Med Chem 2020;191:112107. [Crossref] [PubMed]
- Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019;38:2844-59. [Crossref] [PubMed]
- Li J, Wang R, Jin J, et al. USP7 negatively controls global DNA methylation by attenuating ubiquitinated histone-dependent DNMT1 recruitment. Cell Discov 2020;6:58. [Crossref] [PubMed]
- Yarychkivska O, Tavana O, Gu W, et al. Independent functions of DNMT1 and USP7 at replication foci. Epigenetics Chromatin 2018;11:9. [Crossref] [PubMed]
- Sadeghi H, Esmkhani S, Pirjani R, et al. CREB-binding protein (CREBBP) and preeclampsia: a new promising target gene. Mol Biol Rep 2021;48:2117-22. [Crossref] [PubMed]
- Chen X, Wang K, Jiang S, et al. eEF2K promotes PD-L1 stabilization through inactivating GSK3β in melanoma. J Immunother Cancer 2022;10:e004026. [Crossref] [PubMed]
- Hu X, Li J, Fu M, et al. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther 2021;6:402. [Crossref] [PubMed]
- Xin P, Xu X, Deng C, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol 2020;80:106210. [Crossref] [PubMed]
- Liu J, Zhang C, Wang J, et al. The Regulation of Ferroptosis by Tumor Suppressor p53 and its Pathway. Int J Mol Sci 2020;21:8387. [Crossref] [PubMed]
- Duchartre Y, Kim YM, Kahn M. The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol 2016;99:141-9. [Crossref] [PubMed]
- Liu G, Chen T, Ding Z, et al. Inhibition of FGF-FGFR and VEGF-VEGFR signalling in cancer treatment. Cell Prolif 2021;54:e13009. [Crossref] [PubMed]
- Moon H, Ro SW. MAPK/ERK Signaling Pathway in Hepatocellular Carcinoma. Cancers (Basel) 2021;13:3026. [Crossref] [PubMed]
- Guo YJ, Pan WW, Liu SB, et al. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med 2020;19:1997-2007. [PubMed]
- Yang D, Zhou Q, Labroska V, et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther 2021;6:7. [Crossref] [PubMed]
- Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer 2007;7:79-94. [Crossref] [PubMed]
- He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res 2020;30:660-9. [Crossref] [PubMed]
- Leclerc M, Voilin E, Gros G, et al. Regulation of antitumour CD8 T-cell immunity and checkpoint blockade immunotherapy by Neuropilin-1. Nat Commun 2019;10:3345. [Crossref] [PubMed]
- Bag A, Ghorai PK. Development of Quantum Chemical Method to Calculate Half Maximal Inhibitory Concentration (IC50). Mol Inform 2016;35:199-206. [Crossref] [PubMed]
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol 2020;72:250-61. [Crossref] [PubMed]
- Yang JD, Heimbach JK. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ 2020;371:m3544. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Xu D, Du M, Zhang J, et al. DNMT1 mediated promoter methylation of GNAO1 in hepatoma carcinoma cells. Gene 2018;665:67-73. [Crossref] [PubMed]
- Huang A, Yang XR, Chung WY, et al. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther 2020;5:146. [Crossref] [PubMed]
- Sangro B, Sarobe P, Hervás-Stubbs S, et al. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:525-43. [Crossref] [PubMed]
- Zhou J, Sun H, Wang Z, et al. Guidelines for the Diagnosis and Treatment of Hepatocellular Carcinoma (2019 Edition). Liver Cancer 2020;9:682-720. [Crossref] [PubMed]
- Craig AJ, von Felden J, Garcia-Lezana T, et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2020;17:139-52. [Crossref] [PubMed]
- Wang X, Zhang Q, Wang Y, et al. Clinical Significance of Ubiquitin Specific Protease 7 (USP7) in Predicting Prognosis of Hepatocellular Carcinoma and its Functional Mechanisms. Med Sci Monit 2018;24:1742-50. [Crossref] [PubMed]
- Yamaguchi L, Nishiyama A, Misaki T, et al. Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation. Sci Rep 2017;7:55. [Crossref] [PubMed]
- Zhu Y, Ye F, Zhou Z, et al. Insights into Conformational Dynamics and Allostery in DNMT1-H3Ub/USP7 Interactions. Molecules 2021;26:5153. [Crossref] [PubMed]
- Zhong Z, Virshup DM. Wnt Signaling and Drug Resistance in Cancer. Mol Pharmacol 2020;97:72-89. [Crossref] [PubMed]


