
Glucocorticoid receptor regulates the epithelial-mesenchymal transition process through GR/ZEB1/E-cad and is involved in breast cancer endocrine drug resistance—a bioinformatics analysis
Highlight box
Key findings
• Glucocorticoid receptor (GR) participates in endocrine resistance by GR/ZEB1/E-cad regulation of epithelial-mesenchymal transition (EMT).
What is known and what is new?
• GR is involved in breast cancer endocrine therapy resistance by regulating the EMT process.
• GR is involved in endocrine resistance to breast cancer through the EMT-related gene, ZEB1. ZEB1 is involved in the EMT process through downstream SETD1B.
What are the implications, and what should change now?
• We investigated the role and mechanism of GR in the EMT process and endocrine resistance of estrogen receptor positive/human epidermal growth factor receptor-2 negative breast cancer.
• This study mostly used data from public databases, but this data may be incomplete, such as the via omission of relevant genes and proteins.
Introduction
Background
According to the latest cancer research data, breast cancer has replaced lung cancer as the most common malignant tumor worldwide (1). About 70% of diagnosed breast cancer patients are estrogen receptor (ER)-positive (ER+). Endocrine therapy that blocks the ER signaling pathway is widely used in the treatment of hormone receptor-positive breast cancer (2). However, about 30% of ER+ patients have primary resistance to endocrine therapy, and almost all advanced patients who respond to initial treatment will develop secondary resistance after a period of treatment (3). The development of endocrine resistance (either de novo or acquired) is a clinically challenging issue in the management of ER+ breast cancer. Although new agents have emerged in recent decades, such as CDK4/6 inhibitors [previous studies have demonstrated that cyclin-dependent kinase 4/6 (CDK4/6) inhibitors combined with endocrine therapy are able to effectively improve the prognosis of hormone receptor positive (HR+)] (4). Also, endogenous exRNAs are also hot topics in the study of drug resistance in tumor cells (5). endocrine resistance remains an unresolved and demanding problem (6).
The response elements of glucocorticoid receptor (GR) and ER have a high degree of DNA overlap (7). Region-specific GR promoter methylation is an independent prognostic marker for patient survival and identifies a subset of patients with poor prognosis, particularly those without tamoxifen treatment (8). Liganded GR can suppress ER chromatin occupancy at shared ER-regulated enhancers, including Cyclin D1 (CCND1), regardless of whether the ligand is a classic GR agonist or antagonist. The resulting GR-mediated suppression of ER+ breast cancer proliferative gene expression and cell division suggests that a selective GR modulator (SGRM) could decrease ER-driven gene expression (9). Previous studies have shown that ER, progesterone receptor (PR), androgen receptor (AR), and GR have similar properties on the promoters of target genes. The binding sequence (a couple of the binding sites) of several repeating sequences such as 5'-AGGTCA and 5'-AGAACA are separated by three nucleotides, as in IR3, an inverted repeat sequence separated by three base pairs (bps), or DR3, a direct repeat sequence separated by three bps (10,11). GR promotes the expression of activating protein-1 (AP1) and forkhead box A1 (FoxA1), which bind to the ER promoter and inhibit the expression of ER, indicating that there may be cross-regulation between the GR and ER pathways. However, GR appears to play opposing roles in ER+ and ER-negative (ER−) breast cancers. Treatment with physiologic concentrations of GC reduces the sensitivity of triple-negative breast cancer (TNBC) to chemotherapy both in vitro and in vivo, and the subsequent administration of a GR antagonist increases the cytotoxicity of paclitaxel, which is not induced by a GR antagonist alone (12,13). In early ER− breast cancer patients, the activation of GR is associated with the expression of a series of genes related to tumor survival and chemotherapy resistance, such as apoptosis inhibition pathways and epithelial-mesenchymal transition (EMT) pathways (7).
Glucocorticoids induce effects that are dependent upon the GR-mediated transcriptional regulation of specific genes known to play key roles in cellular/tissue functions, including growth, apoptosis, differentiation, metastasis and cell survival. Glucocorticoids, like dexamethasone, can increase the expression of c-fms in breast cancer cells in a laboratory setting, through a pathway that depends on the GR. This is associated with an increase in the invasiveness of the cancer. The research suggests that glucocorticoids regulate the growth and differentiation of cancerous breast cells, at least partially, by controlling the activity of the c-fms gene. The c-fms gene encodes a receptor called CSF-1 receptor (CSF-1R), which is a growth factor. Abnormal expression of CSF-1 and CSF-1R is linked to higher risk of disease and poorer outcomes. On the other hand, introducing c-fms into normal breast cells promotes cellular invasion and growth without the need for anchorage. In a mouse model of experimental metastasis, injecting human breast cancer cells with high levels of c-fms into the spleen has resulted in increased tumor growth and spread compared to control breast cancer cells (14). Active RAS proteins activate multiple signaling pathways that regulate various cellular processes such as proliferation, differentiation, migration, metastasis, senescence, and survival. These pathways include RAF-MAPK, PI3K-AKT, RASSF5-MST1/2, TIAM1-RAC, and ASPP1/2-p53. These pathways are crucial for tumor development. Recent evidence suggests that GR influences RAS-dependent signaling and RAS activation. When unliganded, GR reduces RAS activation. However, upon ligand binding, GR dissociates from RAS complexes, moves into the nucleus, and triggers the activation of RAS and its downstream pathways. GR interacts with RAS and RAF1, as well as their associated proteins, such as members of the 14-3-3 family of adapter proteins (15).
A panel of TNBC breast cancer cell lines was treated with cortisol, which resulted in epigenetic alteration characterized by the loss of methylation on the promoter regions of tumor suppressor genes including estrogen receptor 1 (ESR1) and the loss of methylation on the long interspersed nuclear elements-1 (LINE-1) repetitive element, which is used as a surrogate marker for global methylation (16,17). Studies have shown that in patients with ER+ breast cancer, high GR levels can improve the recurrence-free survival (RFS) of patients, while low GR levels can shorten the RFS (18,19). Mechanistic studies have shown that in ER+/GR+ mouse breast cancer cells, ER and GR can mutually enhance each other’s chromatin openness; at the same time, the high expression of GR can induce ER activation (20,21). In the human luminal breast cancer cell line MCF-7, co-activation of ER and GR increased the expression of genes associated with cell differentiation while inhibiting the Wnt/β-catenin signaling pathway, as compared to ER activation alone (22,23). Moreover, compared with the activation of GR alone, the co-activation of GR and ER can down-regulate the expression of GR-regulated EMT-related genes and increase the phosphorylation level of β-catenin, but the specific mechanism remains unknown. Endocrine resistance to ERα confers increased invasive proliferation and induced cell proliferation to epithelial cells. ERα signaling may affect transcription factors that govern EMT. Knockout or silence the ERα and ERβ in MCF-7 breast cancer cells, respectively, lead to changes in phenotype, cellular function, transcription and protein levels of EMT epithelial markers (21). ER binding sites are not found on the target gene promoters of core transcription factors of EMT, such as Snail, Slug, zinc finger E-box binding homeobox 1 (ZEB1), and ZEB2, but there are GR binding sites on them, accounting for the largest number among steroid receptors (24). There is evidence that breast cancer endocrine therapy resistance may be related to EMT-related genes, such as Slug, Snail, Twist, and ZEB (12).
Our theory states that GR is connected to a gene involved in the EMT process via a gene or a pathway, and this connection is related to the endocrine resistance of breast cancer. To identify associated genes, we used bioinformatics analysis for gene enrichment analysis and retrograde correlation analysis to draw scientific conclusions. The majority of current research on the sex hormone receptors ER, PR, AR, PI3K pathway, MAPK pathway, cell cycle, and other intra- and extracellular signaling pathways is focused on the resistance of breast cancer to endocrine therapy. A significant problem in the treatment of breast cancer is figuring out the regulatory processes that lead to resistance to endocrine therapy and developing strategies to overcome them. This study aimed to confirm the role of GR and EMT in ER+/Her2− breast cancer and provide favorable evidence supporting GR as a new target of endocrine drug resistance.
Rationale and knowledge gap
GR, ER, PR, and AR all belong to the steroid receptor family and have similar structures, including a hormone-binding region, a DNA-binding region, and a nuclear transcription region. Different steroid receptors interact in the promoter region of target genes due to the nuclear co-localization mechanism. It has been shown that there is a cross-regulation between ER and GR, which can affect the expression of GR-regulated EMT genes. We have previously found that GR expression was significantly upregulated in recurrent or metastatic foci of endocrine-resistant breast cancer patients, and the expression of the epithelial-like cell marker E-cad was significantly downregulated, while that of the mesenchymal-like cell marker Vimentin was upregulated, suggesting the mesenchymal transformation of tumors. We hypothesized that the suppression of ER activation during endocrine therapy mediates the upregulation of GR-regulated EMT-related gene expression through Wnt/β-catenin, leading to EMT in breast cancer cells. However, the exact mechanism involved is unclear.
Objective
This study will explore possible molecular mechanisms of endocrine therapy resistance in ER+/Her2− breast cancer and attempt to identify any targets for this pathway. We present this article in accordance with the STREGA reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1628/rc).
Methods
In step 1, we screened and analyzed genes to find the ones that are expressed differently. We repeated this process in step 2. In step 3, we performed enrichment analysis on these genes using GO and KEGG. For step 4, we created a PPI network and used Cytoscape to find important genes. In step 5, we focused on finding key proteins associated with specific pathways that have a strong correlation with the important genes. Finally, in step 6, we conducted functional identification and correlation analysis of these key proteins to understand their role in endocrine resistance (Figure 1).
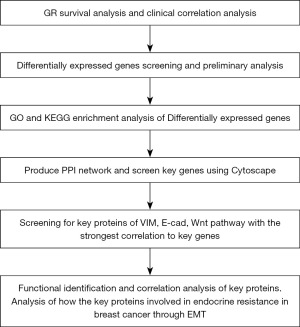
Data acquisition and processing
We obtained ER+/Her2− breast cancer data from METABRIC (https://www.cbioportal.org/study/summary?id=brca_metabric), used the impute package for data cleaning and gap-filling, merged the NR3C1 [NR3C1 encodes GRs, is located on chromosome 5 (region 5q31.3) (25)] gene expression data with ER+/Her2− breast cancer patient survival data, and obtained 1,475 gene identity document (ID) numbers. We also obtained a preprocessed dataset (GSE139870) from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database, which was established based on the anti-proliferative transcriptional effects of medroxyprogesterone acetate in ER+ breast cancer cells predominantly mediated by the progesterone receptor (HG-U133A) (26). This dataset contains three low GR expression samples and three high expression samples of the ER+/Her2− breast cancer cell lines. Moreover, we obtained all of the related proteins of Wnt/β-catenin, E-Cad, and Vimentin from TCGA (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and screened them using P<0.05 and correlation >0.2 (Pearson test), finally obtaining a total of 18,303 proteins. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
GR correlation analysis
We obtained the ER+/Her2− breast cancer data from METABRIC, merged the NR3C1 (GR) gene expression data with the survival data of ER+/Her2− breast cancer patients, drew the survival curve by binary classification of NR3C1 (GR) expression, and observed the survival relationship between the two curves. Survival analysis was performed using the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service) to verify the OS of ER+/Her2−. A total of 603 patients were screened to verify the NR3C1 gene. Clinical correlation analysis was performed in UALCAN (http://ualcan.path.uab.edu/analysis.html) to observe the expression of NR3C1 in different types of breast cancer (normal, luminal, HER2, TNBC).
Next, we obtained the uniform standardized pan-cancer dataset, TCGA Pan-Cancer (PANCAN, N=10,535), from the University of California Santa Cruz Genome Database (UCSC) (https://genome.ucsc.edu), from which we further extracted the ENSG00000139718 (SETD1B) gene expression data in each sample (27). We further screened the samples from solid tissue normal, primary blood-derived cancer-peripheral blood, and primary tumor, and performed log2 transformation for each expression value. Finally, we also eliminated cancer types with less than three samples and obtained the expression data for 26 types of cancer.
Screening of differentially expressed genes
We used the limma package for data preprocessing, setting P<0.05 for screening, and obtained a gene expression matrix containing a total of 1,767 genes. We then used the ggplot2, clusterProfiler, and heatmap packages and classified the original data according to the GR expression level to construct volcano maps and heat maps (28,29).
Functional enrichment and pathway analysis of differentially expressed genes
We used the ggplot2 and clusterProfiler packages to perform GO and KEGG enrichment analysis on the differentially expressed genes, selecting P<0.05 as the cutoff value. We also identified the biological processes (bps), cellular components (CCs), and molecular functions (MFs) of the differential genes. Next, we performed a KEGG pathway enrichment analysis of the differential genes to analyze the biological metabolic and identified pathways of differential gene enrichment.
PPI network construction and screening of core genes
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) was used to evaluate PPIs in the functional protein association networks. A PPI network of differentially expressed genes was constructed using the STRING database. The node genes downloaded from the PPI network were screened according to logFC >2 (log fold change) or logFC <−2. Visualization was performed to hide the unconnected genes by Cytoscape (30). Molecular Complex Detection (MCODE) for Cytoscape to find gene functional clusters with a degree cutoff =2, Node Score Cutoff =0.2, k-core =2, and Max Depth =100 in the PPI network. Then, we filtered out the modules with scores >5 and employed CytoHubba in Cytoscape to identify the top 10 key genes (31).
Screening key proteins in Wnt/β-catenin, E-Cad, Vimentin
We obtained the E-Cad, Vimentin, and Wnt/β-catenin pathway-related proteins in breast cancer from TCGA database, set P<0.05 and correlation >0.15 for screening, and then analyzed the co-expression of the key genes and all proteins. According to the analysis results, we screened out two proteins that are both related to the core genes and have a high degree of correlation: ZEB1 and GLI family zinc finger 2 (GLI2).
Clinical data demonstration
We used UALCAN for further clinical correlation analysis. We used METABRIC to download the processed ER+/Her2− breast cancer survival data, GR expression data, and ZEB1 and GLI2 expression data. Next, we drew a receiver operating characteristic (ROC) curve.
Statistical analysis
All statistical analyses were conducted using R (version 4.3). We considered P values less than 0.05 as statistically significant, using a two-sided approach. Descriptive statistics were employed to analyze data from ER-positive/HER-2-negative breast cancer patients in TCGA, Metabric, and TCGA. Categorical variables were presented using frequencies and proportions, while continuous variables were reported as means plus standard deviations.
Results
Validation of GR’s clinical relevance
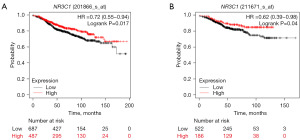
A positive correlation between high GR levels and longer OS was identified on analysis of NR3C1 expression. On analysis with the Kaplan-Meier Plotter and UALCAN databases, 2,241 patients with ER+/Her2− receiving endocrine therapy and chemotherapy. The conclusions obtained were similar as those from the METABRIC database, with a positive correlation between high GR levels and longer OS.
Using UALCAN for clinical correlation analysis, we found that the GR expression was highest in patients with luminal-type breast cancer, as compared to those with HER2 and TNBC breast cancer (Figure 2). To further investigate the correlation between GR and the efficacy of endocrine therapy for ER+/Her2− breast cancer, we performed survival analysis using the Kaplan-Meier Plotter database for patients with ER+/HER2− breast cancer treated with endocrine therapy and screened the pathological staging of ER+/Her2− patients treated with endocrine therapy. In total, 1,174 patients were analyzed for RFS survival and 708 patients for DMFS (distant metastasis-free survival), and the results again showed that high GR levels were positively correlated with longer RFS and DMFS (Figure 3).

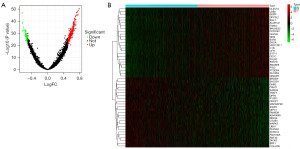
Differentially expressed genes screening and preliminary analysis
We created volcano plots and heat maps to compare the differentially expressed genes between the low-expression group (t) and the high-expression group (n) screened from the GEO database. Analysis of the differentially expressed genes was based on the condition of P<0.05; a total of 1,767 differential genes were finally screened out and a heat map generated (Figure 4).
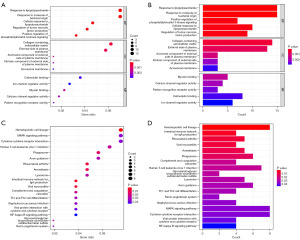
GO and KEGG enrichment analysis of differentially expressed genes
We also performed GO and KEGG enrichment analyses to study the function of the differentially expressed genes. The ggplot2 and clusterProfiler packages were used to determine the enriched differential genes in terms of BP, CC, and MF, respectively. The differentially expressed genes were mainly enriched in the extracellular matrix, tumor necrosis factor signaling pathway, and PI3K signaling pathway (Figure 5). The down-regulated and up-regulated genes were subjected to KEGG enrichment analysis, respectively, and the results showed that the down-regulated genes were mainly enriched in neuroactive ligand-receptor interactions, the PI3K pathway, cell adhesion molecules, and cytokine-cytokine receptor interactions; while the up-regulated genes were mainly enriched in cancer microRNAs including breast cancer, immune-related pathways (Figure 6).

Establishing a PPI network and screening for key genes
A PPI network was constructed using the STRING bioinformatics database to explore the interaction between the proteins corresponding to the differentially expressed genes. The node genes obtained from the PPI network were screened according to logFC>2 or logFC<−2, and 78 differentially expressed genes were identified. Cytoscape software was employed to visualize the entire PPI network. The PPI network was drawn using node and logFC, consisting of 274 nodes and 647 edges. Cytoscape was also applied to construct a protein interaction network, and we introduced betweenness centrality (BC) to draw the graphics (Figure 7). The MCODE plugin in Cytoscape was used to build the functional modules; the results showed that there were three modules with a specific score >6.

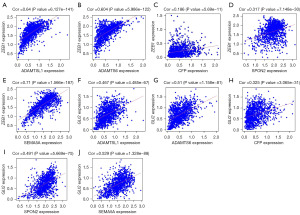
The top 10 key genes were obtained by filtering and intersecting with the genes of the core module using the topology method of the CytoHubba plugin in Cytoscape. The key genes were as follows: CFP, SEMA5A, ADAMTS15, ADAMTSL1, ADAMTSL4, THSD7B, ADAMTS6, and ADAMTS13 (Figure 8). ADAMTS6 suppresses tumor progression via the ERK (extracellular regulated protein kinases) signaling pathway and serves as a prognostic marker in human breast cancer (18). Germline variation in ADAMTSL1 is associated with prognosis following breast cancer treatment in young women (19). ADAMTSL1 and ADAMTS6 were found to be associated with ER+/Her2− breast cancer; the CFP, SPON2, and SEMA5A genes had the highest core scores; and finally, five genes (ADAMTSL1, ADAMTS6, CFP, SPON2, and SEMA5A) were screened as key genes.

Screening for key proteins
To investigate how the GR key genes participate in the EMT process of ER+/HER2− breast cancer by regulating the epithelial marker E-cad, the mesenchymal marker Vimentin, and the core pathway Wnt/β-catenin during EMT, we downloaded all related proteins of Wnt/β-catenin, E-Cad, and Vimentin in breast cancer from TCGA, set P<0.05 and correlation degree >0.15 for screening, and obtained a total of 18,303 proteins. We then correlated the five GR key genes with these 18,303 proteins and found that the ZEB1 and GLI2 proteins were related to the five key genes and were highly correlated according to the analysis (Figure 9). ZEB1 and GLI2 are key proteins involved in the EMT process, and thus, we speculated that GR is involved in ER+/Her2− breast cancer EMT process through ZEB1 or GLI2.
Key protein function validation and survival analysis
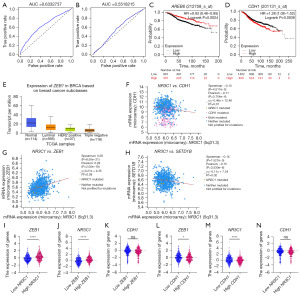
We performed survival and clinical correlation analyses of ZEB1 and GLI2 to demonstrate the function and efficacy of key proteins. In the METABRIC analysis, we screened 1,475 patients aged 30–90 years who were pathologically typed as ER+/Her2− and were receiving endocrine therapy and chemotherapy. By constructing ROC curves, we found that ZEB1 (AUC =0.633) was more clinically relevant than that of GLI2 (AUC =0.552). We considered that ZEB1 is an EMT core transcription factor that could directly repress E-cad expression and participate in the EMT process. Validation using the UALCAN database on 833 breast cancer patients in TCGA revealed that ZEB1 was most expressed in ER+/HER2− breast cancers, as compared to other types of breast cancer.
Further validation using the Kaplan-Meier Plotter database identified 1,174 patients with ER+/Her2− breast cancer who were receiving endocrine therapy. The results showed a positive correlation between high ZEB1 levels and longer RFS in patients with ER+/Her2− breast cancer treated with endocrine therapy, and a negative correlation between high CDH1 levels in ER+/Her2− breast cancer patients receiving endocrine treatment and longer RFS. Also, NR3C1 expression was positively correlated with ZEB1 expression and negatively correlated with CDH1 expression (Figure 10).
ZEB1 participates in the EMT process through its downstream SETD1B
The EMT transcription factor, ZEB1, alters the epigenetic landscape of colorectal cancer cells (20). A positive feedback loop between ZEB1 and its downstream ZEB1 in colorectal cancer has bene identified that maintains the EMT regulatory state. We further analyzed ZEB1, the gene downstream of ZEB1. TCGA showed that the expression of ZEB1 was significantly upregulated in breast cancer, and the cBioPortal showed that the expression of the ZEB1 mutant type was higher than that of the wild type in breast cancer. These results suggest that ZEB1 may be associated with breast cancer. We also used the Kaplan-Meier Plotter database to perform a survival analysis of ZEB1 expression in ER+ breast cancer patients receiving endocrine therapy and screened 1,174 patients with a pathological staging of ER+/Her2− receiving endocrine therapy. The results showed that high ZEB1 levels were positively associated with longer RFS in patients with ER+/Her2− breast cancer treated with endocrine therapy.
Correlation analysis revealed that SETD1B expression was negatively correlated with ZEB1 expression, positively correlated with CDH1 expression, and negatively correlated with NR3C1 expression in ER+ breast cancer treated with endocrine therapy (Figure 11), which is opposite to the role of ZEB1 in colorectal cancer. Considering that ZEB1 plays a contrasting role in ER+ breast cancer relative to colorectal cancer, GR may affect EMT by regulating its downstream ZEB1 through ZEB1.

Discussion
Key findings
GR is involved in endocrine therapy resistance in breast cancer
GR has a similar structure to ER, PR, and AR. Previous studies have shown that the response elements of GR and ER have highly overlapping DNA sequences, and there is cross-regulation between ER and GR (12,13). Research has also found that GR may regulate the EMT process through the Vimentin, E-cad, and Wnt/β-catenin pathway and thus be involved in secretory drug resistance in breast cancer (19). In ER+ early-stage breast cancer patients, the high expression of GR is associated with improved RFS. This is consistent with the positive correlation between high GR levels and longer OS in ER+/Her2− breast cancer patients observed in our study, the improvement of OS in ER+/Her2− breast cancer patients by GR, and the improvement of RFS and DMFS in ER+/Her2− breast cancer patients treated with endocrine therapy by GR.
GR is involved in breast cancer endocrine therapy resistance by regulating the EMT process
Previous studies have shown that the cross-regulation between ER and GR can influence the expression of GR-regulated EMT genes and that GR may be involved in breast cancer endocrine resistance by regulating the EMT process through the mesenchymal marker Vimentin, the epithelial marker E-cad, and the Wnt/β-catenin pathway (18,19). This is consistent with the results of the retrograde correlation analysis between the core genes screened in this study and genes related to the Vimentin, E-cad, and Wnt/β-catenin pathways in ER+/Her2− breast cancers.
GR may mediate endocrine resistance to breast cancer via the EMT-related gene, ZEB1
Previous studies have shown that endocrine therapy resistance in breast cancer is associated with EMT-related genes, such as Slug, Snail, Twist, and ZEB. ZEB1 is overexpressed in breast cancer, regulates cell adhesion and polarity, regulates chemoresistance and radioresistance, and promotes breast cancer stem cell generation (21-24). ZEB1 expression in a miR (microRNA)-200-dependent or miR-200-independent manner leads to E-cad transcriptional silencing and cancer progression.
This is consistent with the results of the retrograde correlation analysis of the GR core gene with Vimentin, E-cad, and Wnt/β-catenin pathway genes in this study. GR and ZEB1 were lowly expressed in ER+ breast cancer. Through further correlation analysis of ZEB1, we found that GR was positively correlated with ZEB1 expression, negatively correlated with ZEB1 expression, a downstream protein of ZEB1, and negatively correlated with E-cad. In contrast, ZEB1 expression was negatively correlated with E-cad and negatively correlated with ZEB1. High ZEB1 levels in ER+/Her2− breast cancer treated with endocrine therapy were positively correlated with longer RFS in ER+/Her2− breast cancer treated with endocrine therapy, and high CDH1 levels were negatively correlated with longer RFS in ER+/Her2− breast cancer treated with endocrine therapy. This is consistent with the speculation that GR may inhibit the expression of ZEB1, via GR/ZEB1. Notably, it is known that ZEB-1 is a transcriptional regulator of epithelial-mesenchymal transformation which translates to the more malignant behavior of cancer and therefore shortened survival. We hypothesize that there may be a negative feedback mechanism between GR and ZEB1 in ER+ breast cancer, and that when ZEB1 is highly expressed, it will suppress GR expression and ultimately inhibit the EMT process, improving patient prognosis. Recently, a study showed that hsa-miR-497-3p controls EMT in TNBC cells by targeting ZEB1. This was proven by the changes in the expression of vimentin, N-cadherin, and E-cadherin due to ZEB1 inhibition. The study suggests that hsa-miR-497-3p can reduce the growth and invasion of TNBC cells by regulating EMT through ZEB1 targeting (32).
ZEB1 may be involved in the EMT process through its downstream gene SETD1B
Previous studies have shown that there is a positive feedback loop between ZEB1 and its downstream gene ZEB1, maintaining the EMT regulatory state in colorectal cancer. This is consistent with our finding that ZEB1 gene expression is correlated with E-cad and ZEB1 in the present study. However, we also found that the negative regulatory relationship between ZEB1 and ZEB1 in ER+ breast cancer is different from the positive feedback regulatory relationship between them in colorectal cancer. Considering that ZEB1 plays an opposite role in ER+ breast cancer that that in colorectal cancer, GR may affect EMT through ZEB1 regulation of its downstream gene ZEB1.
In the tumor microenvironment, ZEB1 has been found to play an increasingly important role. The study has shown that ZEB1 is a key factor in the tumor microenvironment and for maintaining tumor-associated macrophages (TAMs)’ tumor-promoting functions (33). The fibrotic microenvironment promotes the spread of tumor cells to the lungs by mediating the ZEB1-AS1/miR-200b-3p/ZEB1 signaling pathway (34). Removing ZEB1 reduces the ability of breast cancer cells to grow, spread, and change their characteristics. Additionally, we have shown that endothelial cells derived from the tumor microenvironment provide the Notch ligand Jagged1 (Jag1) to neighboring breast cancer stem cells through direct cell-cell contact. This leads to an increase in ZEB1 levels through the Notch1 pathway. In response, tumor cells with ectopic ZEB1 produce more VEGFA and induce Jag1 in endothelial cells in a paracrine manner. Removing ZEB1 disrupts this positive feedback loop in the tumor perivascular niche, which ultimately reduces tumor growth and progression both in vivo and in vitro (35). A study observed that the conditioned medium from ZEB1-expressing MDA-MB-231 cells increased the formation of capillary tubes in human umbilical vein endothelial cells (HUVECs). On the other hand, when ZEB1 was knocked down using RNA interference, it had the opposite effect. ZEB1 also increased the expression of vascular endothelial growth factor A (VEGFA) at both the mRNA and protein levels. When HUVECs were pre-incubated with an anti-VEGFA neutralized antibody, it reduced the tube formation caused by ZEB1. In breast cancer tissues, the expression of ZEB1 was positively correlated with the expression of VEGFA and CD31. At the molecular level, ZEB1 activated the transcription of VEGFA by increasing the recruitment of SP1 to its promoter, which was mediated through the activation of the PI3K and p38 pathways. In a nude mouse xenograft model, study demonstrated that elevated expression of ZEB1 promotes tumorigenesis and angiogenesis in breast cancer (36). ZEB1 has significant potential in tumor microenvironment and endocrine therapy, which brings new opportunities for developing targeted agents.
If the role of the cross-regulation between ER and GR in endocrine therapy resistance of breast cancer is confirmed, GR is expected to become a new target for breast cancer endocrine therapy and will provide a novel treatment idea for endocrine-resistant breast cancer patients.
This study explored the molecular mechanisms of ER+/Her2− endocrine therapy resistance via the effects of GR on EMT. This study is important for identifying potential strategies to combat resistance in ERα+/Her2− breast cancer to endocrine therapy, and suggests a new therapeutic target for improving the clinical prognosis and minimizing medical costs for breast cancer patients.
Since continuous cell and clinical experiments have not been carried out to verify this hypothetical GR/ZEB1/E-cad pathway, more rigorous data supporting this concept is lacking, and cell experiments in ER+/Her2− breast cancer cell line mice will be required in the future. This study mostly used data from public databases, which may be incomplete, such as via the omission of relevant genes and proteins. Exploring the potential mechanism of how GR affects endocrine therapy is crucial for the targeted creation of effective treatment options, enhancing patient resistance to treatment, and extending the duration of time before a relapse occurs.
Implications and actions needed
This study is important for overcoming the common clinical phenomenon of drug resistance in ERα+/Her2− breast cancer endocrine therapy, and suggests a new therapeutic target for improving the clinical prognosis and minimizing medical costs for breast cancer patients. However, more rigorous data supporting this concept is lacking, and cell experiments in ER+/Her2− breast cancer cell line mice will be required in the future.
Conclusions
In this study, we first validated the involvement of GR in endocrine resistance in breast cancer. We screened ZEB1, a key protein of GR associated with the EMT process in ER+/Her2− breast cancer, and found that high ZEB1 levels were positively correlated with longer RFS in ER+/Her2− breast cancer patients receiving endocrine therapy, and high CDH1 levels were negatively correlated with RFS in patients receiving endocrine therapy for ER+/Her2− breast cancer. Also, NR3C1 expression was positively correlated with ZEB1 expression, negatively correlated with CDH1 expression, and negatively correlated with ZEB1, the downstream gene of ZEB1. High ZEB1 levels were negatively correlated with RFS in ER+/Her2− breast cancer receiving endocrine therapy. Furthermore, ZEB1 expression was negatively correlated with ZEB1, while CDH1 expression was positively correlated with CDH1 expression and negatively correlated with NR3C1 expression. The role of ZEB1 in the endocrine resistance of ER+/Her2− breast cancer was verified, further validating the involvement of GR in the endocrine resistance of breast cancer by regulating the EMT process.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1628/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1628/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1628/coif). A.Y. received consulting fees from Daiichi Sankyo and honoraria from Asahi ELLES, AstraZeneca, Chugai, Daiichi Sankyo, Eisai, Eli Lilly, Kyowa Kirin, Nihon Medi-Physics, Pfizer, Taiho and Takeda. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Haque MM, Desai KV. Pathways to Endocrine Therapy Resistance in Breast Cancer. Front Endocrinol (Lausanne) 2019;10:573. [Crossref] [PubMed]
- Harvell DM, Spoelstra NS, Singh M, et al. Molecular signatures of neoadjuvant endocrine therapy for breast cancer: characteristics of response or intrinsic resistance. Breast Cancer Res Treat 2008;112:475-88. [Crossref] [PubMed]
- Zhao S, Zhang H, Yang N, et al. A narrative review about CDK4/6 inhibitors in the setting of drug resistance: updates on biomarkers and therapeutic strategies in breast cancer. Transl Cancer Res 2023;12:1617-34. [Crossref] [PubMed]
- Cao M. The extracellular RNA and drug resistance in cancer: a narrative review. ExRNA 2023;5:1. [Crossref]
- Dimitrakopoulos FI, Kottorou A, Tzezou A. Endocrine resistance and epigenetic reprogramming in estrogen receptor positive breast cancer. Cancer Lett 2021;517:55-65. [Crossref] [PubMed]
- Karmakar S, Jin Y, Nagaich AK. Interaction of glucocorticoid receptor (GR) with estrogen receptor (ER) α and activator protein 1 (AP1) in dexamethasone-mediated interference of ERα activity. J Biol Chem 2013;288:24020-34. [Crossref] [PubMed]
- Snider H, Villavarajan B, Peng Y, et al. Region-specific glucocorticoid receptor promoter methylation has both positive and negative prognostic value in patients with estrogen receptor-positive breast cancer. Clin Epigenetics 2019;11:155. [Crossref] [PubMed]
- Tonsing-Carter E, Hernandez KM, Kim CR, et al. Glucocorticoid receptor modulation decreases ER-positive breast cancer cell proliferation and suppresses wild-type and mutant ER chromatin association. Breast Cancer Res 2019;21:82. [Crossref] [PubMed]
- Mitre-Aguilar IB, Moreno-Mitre D, Melendez-Zajgla J, et al. The Role of Glucocorticoids in Breast Cancer Therapy. Curr Oncol 2022;30:298-314. [Crossref] [PubMed]
- Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Mol Cell Endocrinol 2014;382:97-106. [Crossref] [PubMed]
- Denayer S, Helsen C, Thorrez L, et al. The rules of DNA recognition by the androgen receptor. Mol Endocrinol 2010;24:898-913. [Crossref] [PubMed]
- West DC, Kocherginsky M, Tonsing-Carter EY, et al. Discovery of a Glucocorticoid Receptor (GR) Activity Signature Using Selective GR Antagonism in ER-Negative Breast Cancer. Clin Cancer Res 2018;24:3433-46. [Crossref] [PubMed]
- Moutsatsou P, Papavassiliou AG. The glucocorticoid receptor signalling in breast cancer. J Cell Mol Med 2008;12:145-63. [Crossref] [PubMed]
- Vakili S, Fischer T, Rappaport J. M2 differentiation of MonoMac-1 cell line induced by M-CSF and glucocorticoid pathways. J Cell Physiol 2020;235:7383-91. [Crossref] [PubMed]
- Abduljabbar R, Negm OH, Lai CF, et al. Clinical and biological significance of glucocorticoid receptor (GR) expression in breast cancer. Breast Cancer Res Treat 2015;150:335-46. [Crossref] [PubMed]
- Intabli H, Gee JM, Oesterreich S, et al. Glucocorticoid induced loss of oestrogen receptor alpha gene methylation and restoration of sensitivity to fulvestrant in triple negative breast cancer. Gene 2023;851:147022. [Crossref] [PubMed]
- West DC, Pan D, Tonsing-Carter EY, et al. GR and ER Coactivation Alters the Expression of Differentiation Genes and Associates with Improved ER+ Breast Cancer Outcome. Mol Cancer Res 2016;14:707-19. [Crossref] [PubMed]
- Shi W, Wang D, Yuan X, et al. Glucocorticoid receptor-IRS-1 axis controls EMT and the metastasis of breast cancers. J Mol Cell Biol 2019;11:1042-55. [Crossref] [PubMed]
- Voutsadakis IA. Epithelial-Mesenchymal Transition (EMT) and Regulation of EMT Factors by Steroid Nuclear Receptors in Breast Cancer: A Review and in Silico Investigation. J Clin Med 2016;5:11. [Crossref] [PubMed]
- Sakunrangsit N, Kalpongnukul N, Pisitkun T, et al. Plumbagin Enhances Tamoxifen Sensitivity and Inhibits Tumor Invasion in Endocrine Resistant Breast Cancer through EMT Regulation. Phytother Res 2016;30:1968-77. [Crossref] [PubMed]
- Xie Y, Gou Q, Xie K, et al. ADAMTS6 suppresses tumor progression via the ERK signaling pathway and serves as a prognostic marker in human breast cancer. Oncotarget 2016;7:61273-83. [Crossref] [PubMed]
- Kadalayil L, Khan S, Nevanlinna H, et al. Germline variation in ADAMTSL1 is associated with prognosis following breast cancer treatment in young women. Nat Commun 2017;8:1632. [Crossref] [PubMed]
- Lindner P, Paul S, Eckstein M, et al. EMT transcription factor ZEB1 alters the epigenetic landscape of colorectal cancer cells. Cell Death Dis 2020;11:147. [Crossref] [PubMed]
- Zhou F, Shi Y, Zhao G, et al. A narrative review of the role of glucocorticoid receptors in prostate cancer: developments in last 5 years. Transl Androl Urol 2022;11:1189-99. [Crossref] [PubMed]
- Moore NL, Hanson AR, Ebrahimie E, et al. Anti-proliferative transcriptional effects of medroxyprogesterone acetate in estrogen receptor positive breast cancer cells are predominantly mediated by the progesterone receptor. J Steroid Biochem Mol Biol 2020;199:105548. [Crossref] [PubMed]
- Karolchik D, Baertsch R, Diekhans M, et al. The UCSC Genome Browser Database. Nucleic Acids Res 2003;31:51-4. [Crossref] [PubMed]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 2004;20:3705-6. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Dong Q, Chen H, Li Y, et al. Hsa-miR-497-3p impedes the proliferation and invasion of triple-negative breast cancer cells by controlling epithelial-mesenchymal transition through ZEB1 targeting. Cell Mol Biol (Noisy-le-grand) 2023;69:78-83. [Crossref] [PubMed]
- Cortés M, Sanchez-Moral L, de Barrios O, et al. Tumor-associated macrophages (TAMs) depend on ZEB1 for their cancer-promoting roles. EMBO J 2017;36:3336-55. [Crossref] [PubMed]
- Liu J, Cao L, Meng J, et al. The fibrotic microenvironment promotes the metastatic seeding of tumor cells into the lungs via mediating the ZEB1-AS1/miR-200b-3p/ZEB1 signaling. Cell Cycle 2020;19:2701-19. [Crossref] [PubMed]
- Jiang H, Zhou C, Zhang Z, et al. Jagged1-Notch1-deployed tumor perivascular niche promotes breast cancer stem cell phenotype through Zeb1. Nat Commun 2020;11:5129. [Crossref] [PubMed]
- Liu L, Tong Q, Liu S, et al. ZEB1 Upregulates VEGF Expression and Stimulates Angiogenesis in Breast Cancer. PLoS One 2016;11:e0148774. [Crossref] [PubMed]


