Analysis of prognostic factors and establishment of prediction model of lung adenocarcinoma based on SEER database
Highlight box
Key findings
• In this study, we constructed a nomogram prediction model for lung adenocarcinoma (LUAD) based on the Surveillance, Epidemiology, and End Result database and established a web-based calculator which is easy to use for clinical purposes.
What is known and what is new?
• The nomogram is a statistical predictive model that has been developed for many types of cancer and has shown better performance than traditional tumor, node, and metastasis staging systems.
• There are fewer prediction models to predict survival outcomes in LUAD. The nomogram prediction model developed in this study is important for accurately identifying independent prognostic factors in patients with LUAD and for individualized treatment.
What is the implication, and what should change now?
• The nomogram model constructed in this study has good predictive ability and is expected to be an accurate and personalized tool for predicting the prognosis of LUAD patients.
Introduction
As one of the most frequently diagnosed malignancies, lung cancer is the main cause of cancer-related deaths worldwide, with as estimated 2 million new cases and 1.76 million deaths each year (1). Lung cancer is usually divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC can be further classified as squamous cell carcinoma, lung adenocarcinoma (LUAD), and large cell lung cancer (2). NSCLC accounts for approximately 85% of lung cancer cases and the 5-year overall survival (OS) rate is approximately 15%, LUAD is the most common histological type, accounting for 40% of all lung cancer cases (3,4).
LUAD patients generally have no obvious symptoms in the early stage, and are prone to metastasis and invasion of blood vessels, nerves and lymphatics. Therefore, at the time of diagnosis, two-thirds of patients with LUAD already have advanced stage (IIIB/IV) disease, and the prognosis is poor, with an average 5-year survival rate <20% (5,6). Although therapies for specific tumor mutations, such as anaplastic lymphoma kinase inhibitors, angiogenesis inhibitors, and immunotherapy drugs, can be effective in patients with LUAD, their 5-year OS remains low (7). Therefore, accurate identification of independent factors affecting the prognosis of LUAD patients has important clinical significance for selecting individualized treatment.
The main risk factor for lung cancer is smoking, but among non-smokers, the incidence of lung cancer in women is increasing, especially in Asian (8). A study by Liu et al. found that lung cancer mortality rates declined in the United States from 1990 to 2017, whereas they increased in China, possibly due to different levels of smoking exposure in the U.S. and Chinese populations (9). China continues to face a high burden of lung cancer due to high smoking prevalence and a severely aging population, while the decline in the lung cancer disease burden in the United States has been attributed to the success of smoking cessation campaigns (9,10). Smoking cessation is effective in reducing lung cancer mortality, and public education should continue to be strengthened with a view to the prevention and early detection of lung cancer (9). The growing popularity of chest computed tomography (CT) screening for lung cancer will also affect lung cancer incidence and mortality in China and the United States (11). With this in mind, we used the Surveillance, Epidemiology, and End Results (SEER) database (http://seer.cancer.gov/) to do an analysis of independent risk factors for LUAD patients in the United States and constructed a nomogram prediction model.
The SEER database consists of 18 population-based cancer registries, including nearly 28% of the U.S. population (12). A nomogram is a statistical prediction model that provides a simple graphical representation for use in calculating the numerical probability of clinical events (13). Traditionally, lung cancer staging depends on the tumor, node, and metastasis (TNM) staging system (14). A predictive modeling multivariate nomograms has been developed for many types of cancer and shown be superior to the traditional TNM staging system (15,16). Nevertheless, few nomograms have been used to predict survival outcomes of LUAD.
In this study, we developed a nomogram model to predict cancer-specific survival (CSS) in patients with LUAD and validated the model using an internal validation cohort. The model demonstrated good performance to help clinicians identify individuals at higher risk of cancer-related death and to help personalize treatment program. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-992/rc).
Methods
Patient selection, inclusion criteria and exclusion criteria
All research data were obtained through the SEER*Stat 8.4.0 software. We extracted information from the SEER database on patients diagnosed with lung carcinoma between 2010 and 2015. We defined CSS as the primary endpoint of the study. The following inclusion criteria were applied: (I) patients were diagnosed from 2010 to 2015; (II) patients initially diagnosed with LUAD; (III) the histological type was LUAD (IDO-0-3 codes: 8140); (IV) information on CSS was available. The exclusion criteria were as follows: (I) patients were not initially diagnosed with LUAD; (II) survival data were incomplete; (III) information about the CSS was incomplete; (IV) patients whose detailed information was unknown or unspecified. Figure 1 shows the patient selection criteria and study flow chart. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). No ethics committee approval was required.

Variables selection
In this study, 17 variables relating to the prognosis for each patient were selected. The demographic variables were gender, age, and ethnicity. The cancer characteristics were primary site, laterality, T and N stage, and treatment information (surgery, chemotherapy, and radiotherapy were included). Furthermore, marital status and information on other metastatic sites were extracted. TNM staging was based on the TNM Staging Guidelines (6th edition) published by the International Union for Cancer Control. CSS was defined as the time from medical diagnosis to cancer-related death.
Optimal age cut-off point
Since age was a continuous variable, we used X-tile software to layer the age of the patients to more clearly show the relationship between CSS and different ages (17). Kaplan-Meier method was used for analysis, and the optimal cut-off points were 73 and 80 years. Finally, patients were divided into three age groups for data processing :10–72, 73–79, and 80–99 years (Figure 2).
Statistical analysis
Based on inclusion and exclusion criteria, we eventually included 50,007 patients with LUAD. To establish and validate the nomogram, eligible LUAD patients from the SEER database were divided randomly into the training cohort (70%, n=35,007) and validation cohort (30%, n=15,000) (Tables available at https://cdn.amegroups.cn/static/public/TCR-23-992-1.xls, https://cdn.amegroups.cn/static/public/TCR-23-992-2.xlsx). Univariate Cox regression analysis was used for initial screening of predictor variables. Multivariate Cox regression analysis was used to determine the independent risk factors affecting the prognosis of patients. Based on the results of multivariate analysis, R 4.2.0 (http://www.r-project.org) with “survival” and “rms” packages were used to construct the nomogram (17). The concordance index (C-index) and calibration curve were used to evaluate the performance and accuracy of the nomogram. The C-index ranged from 0.50 to 1.00, with higher values reflecting the greater the reliability of the prediction performance of the model (13). In addition, a reduction of 45 degrees in the diagonal of the calibration curve indicates that this is a perfectly calibrated model. Moreover, area under the curve (AUC) was used to express the discrimination of the nomogram. Data were analyzed using R 4.2.0 software and Statistical Package for the Social Sciences (SPSS) 24.0 software. All confidence interval (CI) were expressed with 95% CI. P<0.05 was considered statistically significance.
Results
Demographic baseline characteristics
A total of 50,007 LUAD patients were randomly separated into the training cohort (n=35,007) and internal validation cohort (n=15,000). The nomogram was then established and validated. The demographic and clinicopathological characteristics of the two cohorts are shown in Table 1. There were no significant differences between the two cohorts in sex, with males and females accounting for 48.27% and 51.73%, respectively. The majority of patients were white and were predominantly in the 10–72 years age group. There were no significant differences between the cohorts in marital status, with 54.77% married and 45.23% unmarried. In terms of the tumor characteristics, the primary site was the upper lobes, the laterality was the unilateral, and the most common T and N stages were T1 (32.93%) and N0 (48.84%). Furthermore, patients with tumor size <3 cm accounted for 45.82%, and lymph node positive accounted for 51.16% of patients. Moreover, metastasis to other sites was not uncommon. There were 9,132 cases (18.26%) of bone metastasis, 6,812 cases (13.62%) of brain metastasis, 3,485 cases (6.97%) of liver metastasis, and 6,143 cases (12.28%) of lung metastasis. In terms of treatment, 35.56% of the patients received surgery, 42.48% received radiotherapy, and 45.76% received chemotherapy.
Table 1
| Variables | Total cohort (n=50,007), n (%) | Training cohort (n=35,007), n (%) | Validation cohort (n=15,000), n (%) | P value | |
|---|---|---|---|---|---|
| Gender | 0.446 | ||||
| Male | 24,140 (48.27) | 16,938 (48.38) | 7,202 (48.01) | ||
| Female | 25,867 (51.73) | 18,069 (51.62) | 7,798 (51.99) | ||
| Age (years) | 0.183 | ||||
| 10–72 | 31,913 (63.82) | 22,357 (63.86) | 9,556 (63.71) | ||
| 73–79 | 10,344 (20.69) | 7,287 (20.82) | 3,057 (20.38) | ||
| 80–99 | 7,750 (15.50) | 5,363 (15.32) | 2,387 (15.91) | ||
| Race | 0.137 | ||||
| White | 39,370 (78.73) | 27,631 (78.93) | 11,739 (78.26) | ||
| Black | 6,598 (13.19) | 4,600 (13.14) | 1,998 (13.32) | ||
| Other | 4,039 (8.08) | 2,776 (7.93) | 1,263 (8.42) | ||
| Marital status | 0.786 | ||||
| Married | 27,390 (54.77) | 19,188 (54.81) | 8,202 (54.68) | ||
| Unmarried | 22,617 (45.23) | 15,819 (45.19) | 6,798 (45.32) | ||
| Primary site | 0.541 | ||||
| Main bronchus | 983 (1.97) | 680 (1.94) | 303 (2.02) | ||
| Upper lobe | 31,421 (62.83) | 21,939 (62.67) | 9,482 (63.21) | ||
| Middle lobe | 2,505 (5.01) | 1,763 (5.04) | 742 (4.95) | ||
| Lower lobe | 14,692 (29.38) | 10,349 (29.56) | 4,343 (28.95) | ||
| Other | 406 (0.81) | 276 (0.79) | 130 (0.87) | ||
| Laterality | 0.794 | ||||
| Unilateral | 49,960 (99.91) | 34,968 (99.89) | 14,992 (99.95) | ||
| Bilateral | 47 (0.09) | 39 (0.11) | 8 (0.05) | ||
| T stage | 0.726 | ||||
| T0 | 20 (0.04) | 16 (0.05) | 4 (0.03) | ||
| T1 | 16,467 (32.93) | 11,486 (32.81) | 4,981 (33.21) | ||
| T2 | 15,557 (31.11) | 10,902 (31.14) | 4,655 (31.03) | ||
| T3 | 9,086 (18.17) | 6,393 (18.26) | 2,693 (17.95) | ||
| T4 | 8,877 (17.75) | 6,210 (17.74) | 2,667 (17.78) | ||
| N stage | 0.435 | ||||
| N0 | 24,425 (48.84) | 17,033 (48.66) | 7,392 (49.28) | ||
| N1 | 4,433 (8.86) | 3,109 (8.88) | 1,324 (8.83) | ||
| N2 | 15,161 (30.32) | 10,687 (30.53) | 4,474 (29.83) | ||
| N3 | 5,988 (11.97) | 4,178 (11.93) | 1,810 (12.07) | ||
| Tumor size (cm) | 0.837 | ||||
| <3 | 22,911 (45.82) | 16,021 (45.77) | 6,890 (45.93) | ||
| 3–5 | 14,543 (29.08) | 10,158 (29.01) | 4,385 (29.23) | ||
| 6–10 | 11,234 (22.46) | 7,901 (22.57) | 3,333 (22.22) | ||
| >10 | 1,319 (2.64) | 927 (2.65) | 392 (2.61) | ||
| Lymph node | 0.201 | ||||
| Negative | 24,425 (48.84) | 17,033 (48.66) | 7,392 (49.28) | ||
| Positive | 25,582 (51.16) | 17,974 (51.34) | 7,608 (50.72) | ||
| Surgery | 0.771 | ||||
| No | 32,227 (64.44) | 22,546 (64.40) | 9,681 (64.54) | ||
| Yes | 17,780 (35.56) | 12,461 (35.60) | 5,319 (35.46) | ||
| Radiation | 0.875 | ||||
| No | 28,764 (57.52) | 20,144 (57.54) | 8,620 (57.47) | ||
| Yes | 21,243 (42.48) | 14,863 (42.46) | 6,380 (42.53) | ||
| Chemotherapy | 0.835 | ||||
| No | 27,125 (54.24) | 18,978 (54.21) | 8,147 (54.31) | ||
| Yes | 22,882 (45.76) | 16,029 (45.79) | 6,853 (45.69) | ||
| Bone metastasis | 0.408 | ||||
| No | 40,875 (81.74) | 28,647 (81.83) | 12,228 (81.52) | ||
| Yes | 9,132 (18.26) | 6,360 (18.17) | 2,772 (18.48) | ||
| Brain metastasis | 0.437 | ||||
| No | 43,195 (86.38) | 30,211 (86.30) | 12,984 (86.56) | ||
| Yes | 6,812 (13.62) | 4,796 (13.70) | 2,016 (13.44) | ||
| Liver metastasis | 0.475 | ||||
| No | 46,522 (93.03) | 32,586 (93.08) | 13,936 (92.91) | ||
| Yes | 3,485 (6.97) | 2,421 (6.92) | 1,064 (7.09) | ||
| Lung metastasis | 0.451 | ||||
| No | 43,864 (87.72) | 30,732 (87.79) | 13,132 (87.55) | ||
| Yes | 6,143 (12.28) | 4,275 (12.21) | 1,868 (12.45) | ||
Analysis of independent prognostic factors in the training cohort
In the training cohort, univariate Cox analysis revealed gender, age, ethnicity, marital status, primary site, laterality, T stage, N stage, tumor size, lymph node, surgery, chemotherapy, radiotherapy, bone metastasis, brain metastasis, liver metastasis, and lung metastasis as independent prognostic factors for CSS (Table 2). In the multivariate Cox regression analysis, gender, ethnicity, age, marital status, N stage, tumor size, surgery, radiotherapy, chemotherapy, bone metastasis, brain metastasis, liver metastasis, and lung metastasis remained as independent prognostic factors for CSS (Table 2).
Table 2
| Variables | Univariate Cox analysis | Multivariate Cox analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Gender | |||||
| Male | Reference | Reference | |||
| Female | 0.746 (0.726–0.767) | <0.001 | 0.773 (0.752–0.795) | <0.001 | |
| Age (year) | |||||
| 10–72 | Reference | Reference | |||
| 73–79 | 1.100 (1.063–1.063) | <0.001 | 1.217 (1.175–1.260) | <0.001 | |
| 80–99 | 1.354 (1.304–1.405) | <0.001 | 1.300 (1.249–1.353) | <0.001 | |
| Race | |||||
| White | Reference | Reference | |||
| Black | 1.075 (1.033–1.118) | <0.001 | 0.956 (0.918–0.996) | <0.05 | |
| Other | 0.899 (0.855–0.947) | <0.001 | 0.701 (0.666–0.738) | <0.001 | |
| Marital status | |||||
| Married | Reference | Reference | |||
| Unmarried | 1.108 (1.078–1.138) | <0.001 | 1.112 (1.081–1.144) | <0.001 | |
| Primary site | |||||
| Main bronchus | Reference | – | – | ||
| Upper lobe | 0.434 (0.399–0.399) | <0.001 | – | – | |
| Middle lobe | 0.425 (0.384–0.384) | <0.001 | – | – | |
| Lower lobe | 0.445 (0.409–0.485) | <0.001 | – | – | |
| Other | 0.530 (0.449–0.626) | <0.001 | – | – | |
| Laterality | |||||
| Unilateral | Reference | – | – | ||
| Bilateral | 1.720 (1.187–2.491) | <0.05 | – | – | |
| T stage | |||||
| T0 | Reference | ||||
| T1 | 0.460 (0.248–0.856) | <0.05 | – | – | |
| T2 | 0.851 (0.458–1.582) | 0.610 | – | – | |
| T3 | 1.280 (0.688–2.381) | 0.435 | – | – | |
| T4 | 1.691 (0.909–3.145) | 0.097 | – | – | |
| N stage | |||||
| N0 | Reference | Reference | |||
| N1 | 1.953 (1.859–2.052) | <0.001 | 1.752 (1.663–1.845) | <0.001 | |
| N2 | 3.200 (3.099–3.304) | <0.001 | 1.968 (1.895–2.044) | <0.001 | |
| N3 | 3.879 (3.723–4.041) | <0.001 | 1.954 (1.863–2.049) | <0.001 | |
| Tumor size (cm) | |||||
| <3 | Reference | Reference | |||
| 3–5 | 3.526 (3.274–3.799) | <0.001 | 1.114 (1.069–1.162) | <0.001 | |
| 6–10 | 1.888 (1.827–1.952) | <0.001 | 1.322 (1.266–1.381) | <0.001 | |
| >10 | 2.831 (2.736–2.929) | <0.001 | 1.590 (1.467–1.722) | <0.001 | |
| Lymph node | |||||
| Negative | Reference | – | – | ||
| Positive | 0.339 (0.320–0.339) | <0.001 | – | – | |
| Surgery | |||||
| No | Reference | Reference | |||
| Yes | 0.192 (0.186–0.199) | <0.001 | 0.273 (0.262–0.286) | <0.001 | |
| Radiation | |||||
| No | Reference | Reference | |||
| Yes | 1.682 (1.636–1.728) | <0.001 | 0.769 (0.746–0.794) | <0.001 | |
| Chemotherapy | |||||
| No | Reference | Reference | |||
| Yes | 1.590 (1.547–1.634) | <0.001 | 0.589 (0.570–0.609) | <0.001 | |
| Bone metastasis | |||||
| No | Reference | Reference | |||
| Yes | 3.479 (3.371–3.590) | <0.001 | 1.714 (1.650–1.781) | <0.001 | |
| Brain metastasis | |||||
| No | Reference | Reference | |||
| Yes | 2.929 (2.830–3.031) | <0.001 | 1.669 (1.612–1.729) | <0.001 | |
| Liver metastasis | |||||
| No | Reference | Reference | |||
| Yes | 3.680 (3.518–3.849) | <0.001 | 1.530 (1.459–1.605) | <0.001 | |
| Lung metastasis | |||||
| No | Reference | Reference | |||
| Yes | 2.577 (2.485–2.671) | <0.001 | 1.081 (1.038–1.125) | <0.001 | |
CI, confidence interval.
Establishment of the nomogram
Finally, we established a nomogram for prediction of CSS in LUAD patients based on the significant independent factors identified in the multivariate Cox regression analysis (Figure 3). The nomogram showed that surgery and chemotherapy shared the largest contribution to prognosis, followed by age and liver metastasis. N stage and tumor size had a moderate impact on survival. Each factor among these variables was assigned a score on the point scale. After locating the total score on the total point scale, the estimated probability of survival at each score point was then estimated by drawing a straight line down to the probability scale. The C-indexes of the training cohort and validation cohort were 0.779 (95% CI: 0.775–0.783) and 0.782 (95% CI: 0.778–0.786), respectively.
Validation and calibration of the nomogram
The receiver operating characteristic (ROC) curve was constructed to evaluate the discrimination ability of the prediction model. The AUC values of the 1-, 3-, and 5-year CSS were 0.700, 0.733, and 0.669 in the training cohort (Figure 4A-4C) and 0.700, 0.744, and 0.669 in the validation cohort (Figure 4D-4F). According to the calibration curve, the observed values of the nomogram showed a strong correlation with the predicted values (Figure 5).


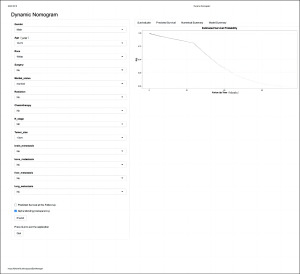
Web-based calculator
Based on the nomogram, we constructed a web-based calculator (https://hjhlovelfb.shinyapps.io/DynNomapp/) to predict CSS rates for each patient by inputting the scores for each of the thirteen variables. The application then provides a survival plot, predicted survival, and numerical summary of the patient by ticking the “Predicted survival at this Follow Up”, “Survival months”, and “Alpha blending (transparency)” options, respectively (Figure 6).
Discussion
Lung cancer is the leading cause of cancer-related mortality (18). LUAD has become the most prevalent lung cancer sub-type, accounting for 50% of all lung cancer diagnoses and its frequency is increasing (19). Patients with LUAD generally have no obvious symptoms in the early stage, and are prone to metastasis and invasion of nerves, lymphatics and blood vessels (20). Patients with LUAD have a poor prognosis, with an average 5-year survival rate of less than 20% (21). Therefore, accurate identification of the independent factors affecting the prognosis of LUAD has important clinical significance for guiding the selection of individualized treatment to improve of efficacy and quality of life of patients.
A study by Woolston et al. demonstrated significant gender and racial differences in the prevalence trends of lung cancer patients (22). Epidermal growth factor receptor (EGFR), which is a transmembrane protein, is one of the most common driver mutations in LUAD (23). In lung cancer, EGFR mutation rates are higher in Asian populations than in Western populations, higher in non-smokers than in smokers, and higher in females than in males (24). A study by Xie et al. based on the SEER database showed that gender was an independent prognostic factor for patients with LUAD, with survival rates being higher in female patients than in males (25). In China, the incidence of LUAD is increasing, and it is common among non-smokers and females.
Campos-Balea et al. (26) identified age, sex, ethnicity and marital status as powerful prognostic variable in patients with LUAD. Multivariate Cox regression analysis confirmed that female sex, age under 65 years and living with others were prognostic factors for OS. TNM stage has also been reported to be a powerful prognostic variable and patients with liver metastasis usually have a high disease burden (27). The most frequent metastatic sites in NSCLC are brain, bone, liver, the respiratory system, and adrenal glands. Furthermore, organ metastases are common indicators of strong tumor invasion, late TNM stage and poor patient prognosis (28,29). Sun et al. (30) reported a negative correlation between tumor diameter and survival time in LUAD patients. Surgical resection is the primary treatment strategy for patients with NSCLC and is often considered the best treatment for lung cancer. Surgical treatment has been identified as an independent factor affecting the prognosis of LUAD patients, presenting a higher survival rate compared with that of non-surgical patients (31). Furthermore, Shi et al. (32) found that chemotherapy significantly prolonged the survival of patients with LUAD.
Won et al. (33) established a nomogram to predict brain metastasis in NSCLC patients based on factors including histological type, N stage, T stage and smoking status. Another nomogram for predicting brain metastasis in NSCLC patients based on factors such as histological type, tumor size, and number of metastatic lymph nodes has also been reported (34). In addition, nomograms have been used to predict the survival of some special types of NSCLC, such as pulmonary invasive mucinous adenocarcinoma. A new nomogram has been established to predict the prognosis of pulmonary invasive mucinous adenocarcinoma based on age, differentiation, TNM stage and treatment (35). Nevertheless, prognostic nomograms for LUAD have not yet been reported.
In this study, we established and validated a nomogram for predicting the CSS rate of LUAD using a large population of patients from the SEER database, and showed that the predictions based on the calibration curves correlated well with the actual observations. In addition, the model includes only important clinically available variables and is less expensive than molecular assays, thus providing economic and practical advantages. Using a variety of statistical methods, we validated the accuracy of the model for predicting the 1-, 3-, and 5-year CSS rates of patients with LUAD using data from the SEER database. Due to the practical limitations associated with the use of paper-based nomograms for predicting CSS rates, web-based calculator has been established to improve the practicability, approachability, and functionality of the prediction model (36,37).
In recent years, with the popularization of low-dose computed tomography (LDCT) in lung cancer screening, the detection rate of pulmonary nodules with subsolid nodules (SSNs) has increased significantly, and it has become an important indicator for lung cancer screening (38,39). The Fleischner Society guidelines recommend an extended period of time before initial follow-up for sub-solid nodules, extending the total length of follow-up to 5 years (40). Computed tomography (CT) scan results and nodule size are important for the treatment and prognosis of early-stage LUAD (41). The development of LUAD risk prediction models not only helps to identify at-risk populations, but also maximizes the impact of LDCT (42). Since the SEER database still lacks data on nodule size and type, the results of this study were mainly used to predict the prognosis of relatively advanced LUAD.
This study has some limitations. First, this study is a retrospective study. Only patients with complete data were included in this study, with inevitable deviations in availability. Second, specific information about systematic treatment, especially details such as the specific types of surgery, dose of radiotherapy and selection of chemotherapy drugs, was not available. Third, the prediction model was not validated with external queues, only with internal queues. Fourth, since the data in this study were from the SEER database, the predictive model we constructed may not be generalized to Chinese patients. However, in the current study, we established a nomogram for the prediction of the prognosis of patients with LUAD based on relevant factors identified by rigorous statistical analysis of a large sample of data. Lastly, as a web-based application, the use of the calculator may be restricted during periods of heavy internet traffic, but this should only be a rare case.
Conclusions
In summary, in this study we developed a predictive model and a web-based calculator to predict individual survival outcomes in LUAD patients effectively and accurately. This model is of great clinical significance for stratifying patients for treatment and promoting the advancement of individualized therapy through the quantitative analysis of survival predictors. Furthermore, this study provides ideas for developing similar clinical prediction models that are not limited to the field of cancer.
Acknowledgments
The authors would like to thank the Surveillance, Epidemiology, and End Results (SEER) database for the support.
Funding: This study was supported by the following grants:
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-992/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-992/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-992/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDeri
References
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Wingo PA, Ries LA, Giovino GA, et al. Annual report to the nation on the status of cancer, 1973-1996, with a special section on lung cancer and tobacco smoking. J Natl Cancer Inst 1999;91:675-90. [Crossref] [PubMed]
- Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 2018;9:117. [Crossref] [PubMed]
- Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet 2011;378:1727-40. [Crossref] [PubMed]
- Lin JJ, Cardarella S, Lydon CA, et al. Five-Year Survival in EGFR-Mutant Metastatic Lung Adenocarcinoma Treated with EGFR-TKIs. J Thorac Oncol 2016;11:556-65. [Crossref] [PubMed]
- Wang G, Xiong R, Wu H, et al. Prognostic Value of Neutrophil-to-lymphocyte Ratio in Patients with Lung Adenocarcinoma Treated with Radical Dissection. Zhongguo Fei Ai Za Zhi 2018;21:588-93. [PubMed]
- Bunn PA Jr, Doebele RC. Genetic testing for lung cancer: reflex versus clinical selection. J Clin Oncol 2011;29:1943-5. [Crossref] [PubMed]
- Wu FZ, Huang YL, Wu CC, et al. Assessment of Selection Criteria for Low-Dose Lung Screening CT Among Asian Ethnic Groups in Taiwan: From Mass Screening to Specific Risk-Based Screening for Non-Smoker Lung Cancer. Clin Lung Cancer 2016;17:e45-56. Erratum in: Clin Lung Cancer 2020;21:e238. [Crossref] [PubMed]
- Liu X, Yu Y, Wang M, et al. The mortality of lung cancer attributable to smoking among adults in China and the United States during 1990-2017. Cancer Commun (Lond) 2020;40:611-9. [Crossref] [PubMed]
- Yang D, Liu Y, Bai C, et al. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett 2020;468:82-7. [Crossref] [PubMed]
- Yotsukura M, Asamura H, Motoi N, et al. Long-Term Prognosis of Patients With Resected Adenocarcinoma In Situ and Minimally Invasive Adenocarcinoma of the Lung. J Thorac Oncol 2021;16:1312-20. [Crossref] [PubMed]
- Zeng Y, Mayne N, Yang CJ, et al. A Nomogram for Predicting Cancer-Specific Survival of TNM 8th Edition Stage I Non-small-cell Lung Cancer. Ann Surg Oncol 2019;26:2053-62.
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Carter BW, Lichtenberger JP 3rd, Benveniste MK, et al. Revisions to the TNM Staging of Lung Cancer: Rationale, Significance, and Clinical Application. Radiographics 2018;38:374-91. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Zaak D, Burger M, Otto W, et al. Predicting individual outcomes after radical cystectomy: an external validation of current nomograms. BJU Int 2010;106:342-8. [Crossref] [PubMed]
- Regression Modeling Strategies, R Package version3.4-0. Available online: http://www.rproject.org/
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Succony L, Rassl DM, Barker AP, et al. Adenocarcinoma spectrum lesions of the lung: Detection, pathology and treatment strategies. Cancer Treat Rev 2021;99:102237. [Crossref] [PubMed]
- Selvaraj G, Kaliamurthi S, Kaushik AC, et al. Identification of target gene and prognostic evaluation for lung adenocarcinoma using gene expression meta-analysis, network analysis and neural network algorithms. J Biomed Inform 2018;86:120-34. [Crossref] [PubMed]
- Chen Y, Xia L, Peng Y, et al. Development and validation of a m(6)A -regulated prognostic signature in lung adenocarcinoma. Front Oncol 2022;12:947808. [Crossref] [PubMed]
- Woolston A, Sintupisut N, Lu TP, et al. Putative effectors for prognosis in lung adenocarcinoma are ethnic and gender specific. Oncotarget 2015;6:19483-99. [Crossref] [PubMed]
- Shi H, Seegobin K, Heng F, et al. Genomic landscape of lung adenocarcinomas in different races. Front Oncol 2022;12:946625. [Crossref] [PubMed]
- Seow WJ, Matsuo K, Hsiung CA, et al. Association between GWAS-identified lung adenocarcinoma susceptibility loci and EGFR mutations in never-smoking Asian women, and comparison with findings from Western populations. Hum Mol Genet 2017;26:454-65. [PubMed]
- Xie B, Chen X, Deng Q, et al. Development and Validation of a Prognostic Nomogram for Lung Adenocarcinoma: A Population-Based Study. J Healthc Eng 2022;2022:5698582. [Crossref] [PubMed]
- Campos-Balea B, de Castro Carpeño J, Massutí B, et al. Prognostic factors for survival in patients with metastatic lung adenocarcinoma: An analysis of the SEER database. Thorac Cancer 2020;11:3357-64. [Crossref] [PubMed]
- Riihimäki M, Hemminki A, Fallah M, et al. Metastatic sites and survival in lung cancer. Lung Cancer 2014;86:78-84. [Crossref] [PubMed]
- Nakazawa K, Kurishima K, Tamura T, et al. Specific organ metastases and survival in small cell lung cancer. Oncol Lett 2012;4:617-20. [Crossref] [PubMed]
- Tamura T, Kurishima K, Nakazawa K, et al. Specific organ metastases and survival in metastatic non-small-cell lung cancer. Mol Clin Oncol 2015;3:217-21. [Crossref] [PubMed]
- Sun F, Ma K, Yang X, et al. A nomogram to predict prognosis after surgery in early stage non-small cell lung cancer in elderly patients. Int J Surg 2017;42:11-6. [Crossref] [PubMed]
- Li F, Zhao Y, Yuan L, et al. Oncologic outcomes of segmentectomy vs lobectomy in pathologic stage IA (≤2 cm) invasive lung adenocarcinoma: A population-based study. J Surg Oncol 2020;121:1132-9. [Crossref] [PubMed]
- Shi M, Zhan C, Shi J, et al. Prediction of Overall Survival of Patients with Completely Resected Non-Small Cell Lung Cancer: Analyses of Preoperative Spirometry, Preoperative Blood Tests, and Other Clinicopathological Data. Cancer Manag Res 2019;11:10487-97. [Crossref] [PubMed]
- Won YW, Joo J, Yun T, et al. A nomogram to predict brain metastasis as the first relapse in curatively resected non-small cell lung cancer patients. Lung Cancer 2015;88:201-7. [Crossref] [PubMed]
- Zhang F, Zheng W, Ying L, et al. A Nomogram to Predict Brain Metastases of Resected Non-Small Cell Lung Cancer Patients. Ann Surg Oncol 2016;23:3033-9. [Crossref] [PubMed]
- Wang Y, Liu J, Huang C, et al. Development and validation of a nomogram for predicting survival of pulmonary invasive mucinous adenocarcinoma based on surveillance, epidemiology, and end results (SEER) database. BMC Cancer 2021;21:148. [Crossref] [PubMed]
- Grimes DA. The nomogram epidemic: resurgence of a medical relic. Ann Intern Med 2008;149:273-5. [Crossref] [PubMed]
- Irish WD, Ilsley JN, Schnitzler MA, et al. A risk prediction model for delayed graft function in the current era of deceased donor renal transplantation. Am J Transplant 2010;10:2279-86. [Crossref] [PubMed]
- Chen PA, Huang EP, Shih LY, et al. Qualitative CT Criterion for Subsolid Nodule Subclassification: Improving Interobserver Agreement and Pathologic Correlation in the Adenocarcinoma Spectrum. Acad Radiol 2018;25:1439-45. [Crossref] [PubMed]
- Li J, Yang X, Xia T, et al. Stage I synchronous multiple primary non-small cell lung cancer: CT findings and the effect of TNM staging with the 7th and 8th editions on prognosis. J Thorac Dis 2017;9:5335-44.
- Tang EK, Chen CS, Wu CC, et al. Natural History of Persistent Pulmonary Subsolid Nodules: Long-Term Observation of Different Interval Growth. Heart Lung Circ 2019;28:1747-54. [Crossref] [PubMed]
- MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. [Crossref] [PubMed]
- Wu FZ, Huang YL, Wu YJ, et al. Prognostic effect of implementation of the mass low-dose computed tomography lung cancer screening program: a hospital-based cohort study. Eur J Cancer Prev 2020;29:445-51. [Crossref] [PubMed]