
Construction of a diagnostic nomogram model for predicting the risk of lymph node metastasis in clinical T1 or T2 colon cancer based on the SEER database
Highlight box
Key findings
• Age, histological grade, histological type, T classification, M classification, and tumour location were independent risk factors identified through univariate and multivariate analyses.
What is known and what is new?
• The incidence of lymph node metastasis (LNM) in patients with pathologic T-stage (pT)1 and pT2 colon cancer was 17.11% according to our study, which has usually been underestimated in the past.
• The nomogram developed in our study can objectively and accurately predict the individual risk of LNM in patients with clinically staged T1 or T2 colon cancer.
What is the implication, and what should change now?
• It is important to assess risk factors to evaluate the presence of LNM, thereby deciding the optimal treatment before further therapy, such as endoscopic resection, which may improve the quality of life in patients with clinically staged T1 or T2 colon cancer rather than radical operation.
Introduction
Background
More than 1.8 million new cases of colorectal cancer and 881,000 deaths from colorectal cancer were expected to occur in 2018, which accounted for almost one-tenth of the total number of new cancer cases and deaths that year. Overall, colorectal cancer ranked third in the incidence rate and second in the death rate of cancer cases (1). A study has shown that poor prognosis and frequent recurrence of colon cancer may be associated with lymph node metastasis (LNM) (2). Moreover, according to the seventh edition of the American Joint Committee on Cancer (AJCC) staging system, when LNM occurs, it is classified as progressive colon cancer (stage III) regardless of the pathologic T-stage (pT) (3).
Rationale and knowledge gap
Statistics have shown that the 5-year relative survival rate is 90% when colon cancer is still in its primary stage, while it decreases to 71% once the patient develops regional metastasis (4). As reported in preceding studies, about 7–16% of T1 colon cancers have been found with LNM (5), and about 18–24% of patients with T2 rectal cancer experience LNM according to a recent study (6). Although radical operation has been widely regarded as the standard treatment for colorectal cancer, some patients with T1 or T2 cancers may be cured by endoscopic dissection after comprehensive evaluation (7). According to the results of the ACOSOG Z6041 study, neoadjuvant chemoradiotherapy followed by local excision might be considered as an organ-preserving alternative in carefully selected patients with clinically staged T2N0 distal rectal cancer who refuse, or are not candidates for, transabdominal resection (8). Beyond that, with regards to patients with rectal cancer, local or endoscopic dissection can help to reduce complications, such as intestinal and sexual dysfunction which may be caused by radical operations, as well as reducing medical costs and shortening hospitalization time. Hence, early identification or prediction of LNM in colon cancer is important for improving treatment strategies as it is a crucial factor for clinically staged T1 or T2 patients to decide whether to proceed with radical surgical treatment. Thus far, there have been numerous models to predict the risk and prognosis of different malignancies (9-12). However, there are few reports or methods related to predicting LNM in patients with clinically staged T1 or T2 colon cancer, and relatively little is known about clinicopathologic risk factors associated with LNM in these patients.
Objective
The nomogram is a widely used and reliable graphical computational model that enables accurate calculation and prediction of individual risk events by considering all the risk factors that affect tumour progression (13). In this study, a nomogram was developed based on data from the Surveillance, Epidemiology, and End Results (SEER) database for predicting the risk of LNM in clinical T1 or T2 colon cancer before further therapy strategies. The purpose was to identify patients at high risk of LNM, which was expected to provide a reference for improving treatment strategies in actual clinical practice. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1451/rc).
Methods
Study population and inclusion and exclusion criteria
Information on patients who developed colon cancer from 2004 to 2015 was retrieved from the SEER database using SEER*Stat 8.4.0.1 software based on the following primary site codes: C18.0 (cecum), C18.2 (ascending colon), C18.3 (hepatic flexure), C18.4 (transverse colon), C18.5 (splenic flexure), C18.6 (descending colon), and C18.7 (sigmoid colon). The following SEER histology codes were used: 8140–8147, 8210–8211, 8220–8221, 8260–8263, 8480–8481, and 8490. The information gathered for each patient consisted of age, race, sex, pathologic type, histologic grade, 6th AJCC stages (T, N, and M stages), tumour location, tumour size, and radiotherapy and chemotherapy information. The inclusion criteria were as follows: (I) patients with primary colon cancer confirmed by microscopic analysis; (II) patients staged as pT1 or pT2 using the sixth edition of the AJCC staging system; (III) patients with colon cancer as the only primary cancer; and (IV) patients who underwent surgical treatment and were subjected to a postoperative examination of complete pathologic specimens. The exclusion criteria were as follows: (I) patients with multiple primary tumours; (II) patients whose case reports came only in the form of an autopsy report or death certificate; (III) patients with missing or incomplete data on one or more of the inclusion criteria; (IV) patients younger than 18 years; and (V) patients who had received neoadjuvant radiotherapy. Based on the above criteria, 32,803 patients were included in this study. They were randomized at a 7:3 ratio into a training set of 22,962 patients and a validation set of 9,841 patients (Table 1). The population in the validation set was used to validate the established nomogram internally. This study was based on publicly available data in the SEER database, and all patient data had been de-identified before collection, so neither ethics committee approval nor informed patient consent was required. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Characteristics | Training set (n=22,962) | Validation set (n=9,841) | χ2 | P |
|---|---|---|---|---|
| Age, n (%) | 4.3818 | 0.1118 | ||
| <45 years | 793 (3.5) | 372 (3.8) | ||
| 45–65 years | 8,003 (34.9) | 3,502 (35.6) | ||
| >65 years | 14,166 (61.7) | 5,967 (60.6) | ||
| Sex, n (%) | 0.20458 | 0.651 | ||
| Male | 10,956 (47.7) | 4,723 (48.0) | ||
| Female | 12,006 (52.3) | 5,118 (52.0) | ||
| Race, n (%) | 3.1759 | 0.3653 | ||
| American Indian/Alaska Native | 142 (0.6) | 49 (0.5) | ||
| Asian or Pacific Islander | 1,783 (7.8) | 798 (8.1) | ||
| Black | 2,764 (12.0) | 1,158 (11.8) | ||
| White | 18,273 (79.6) | 7,836 (79.6) | ||
| Histological grade, n (%) | 0.60925 | 0.8943 | ||
| Grade I | 3,475 (15.1) | 1,504 (15.3) | ||
| Grade II | 17,210 (74.9) | 7,385 (75.0) | ||
| Grade III | 2,034 (8.9) | 847 (8.6) | ||
| Grade IV | 243 (1.1) | 105 (1.1) | ||
| Histological type, n (%) | 5.1255 | 0.07709 | ||
| Adenocarcinoma | 21,472 (93.5) | 9,266 (94.2) | ||
| Mucinous adenocarcinoma | 1,420 (6.2) | 551 (5.6) | ||
| Signet-ring cell carcinoma | 70 (0.3) | 24 (0.2) | ||
| T classification, n (%) | 0.25876 | 0.611 | ||
| T1 | 8,638 (37.6) | 3,732 (37.9) | ||
| T2 | 14,324 (62.4) | 6,109 (62.1) | ||
| N classification, n (%) | 0.50588 | 0.7765 | ||
| N0 | 19,052 (83.0) | 8,137 (82.7) | ||
| N1 | 3,233 (14.1) | 1,415 (14.4) | ||
| N2 | 677 (2.9) | 289 (2.9) | ||
| M classification, n (%) | 0.18531 | 0.6669 | ||
| M0 | 22,509 (98.0) | 9,639 (97.9) | ||
| M1 | 453 (2.0) | 202 (2.1) | ||
| Tumor size, n (%) | 0.019072 | 0.8902 | ||
| <50 mm | 19,538 (85.1) | 8,367 (85.0) | ||
| ≥50 mm | 3,424 (14.9) | 1,474 (15.0) | ||
| Tumor location, n (%) | 1.6982 | 0.9453 | ||
| Ascending colon | 5,078 (22.1) | 2,180 (22.2) | ||
| Cecum | 6,049 (26.3) | 2,582 (26.2) | ||
| Descending colon | 1,176 (5.1) | 506 (5.1) | ||
| Hepatic flexure | 1,001 (4.4) | 428 (4.3) | ||
| Sigmoid colon | 7,083 (30.8) | 3,056 (31.1) | ||
| Splenic flexure | 583 (2.5) | 227 (2.3) | ||
| Transverse colon | 1,992 (8.7) | 862 (8.8) |
Predictive modelling
A prediction model was built based on the independent risk factors and the training set using the “glm” function in R, and then the model was graphically presented as a nomogram using the “regplot” function.
Statistical analysis
Analyses were performed using R software (version 4.1.1). Differences in categorical variables were evaluated using the Chi-squared test. Factors with statistically significant differences (P<0.05) in univariate analysis were included in multivariate logistic regression analysis to identify independent risk factors for LNM. This nomogram was used to assess the individual risk of LNM, and the actual discriminatory power of the nomogram was assessed using the area under the curve (AUC) of the receiver operating characteristic (ROC). The accuracy and clinical utility of the predictive nomogram were assessed using calibration curve analysis and decision curve analysis (DCA), respectively. Statistical significance in the differences was set at P<0.05.
Results
Baseline characteristics of the study population
Of the 32,803 patients who were staged as pT1 or pT2 included for analysis, 5,614 (17.11%) presented with LNMs, while 27,189 (82.89%) did not. There were no significant differences between the training and validation sets in the following baseline characteristics: age, sex, race, histological type, histological grade, T stage, N stage, M stage, tumour size, and tumour location (Table 1), as confirmed by chi-squared test (all P>0.05). Further analysis showed that among the 22,962 patients in the training set, 19,052 were at N0 stage, 3,233 were at N1 stage, and 677 were at N2 stage. Among the 9,841 patients in the validation set, 8,137 were at N0 stage, 1,415 were at N1 stage, and 289 were at N2 stage. The LNM rate was 17% in the training set and 17.3% in the validation set, and the difference was not statistically significant (P=0.5373>0.05).
Univariate and multivariate analyses of risk factors for LNM in early-stage colon cancer
Univariate logistic analysis showed that age, histological grade, histological type, T stage, M stage, tumour size, and tumour location were associated with LNM in patients with colon cancer (P<0.05). By contrast, sex and race were not associated with LNM (P>0.05) (Table 2). The significant indicators identified in the univariate analysis were further included in the multivariate analysis, and the results showed that age, histological grade, histological type, T stage, M stage, and tumour location were all independent risk factors for LNM in early-stage colon cancer (P<0.05) (Table 2).
Table 2
| Characteristics | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | ||
| Age | |||||||
| <45 years | Reference | Reference | |||||
| 45–65 years | 0.658 | 0.588–0.739 | <0.001 | 0.747 | 0.663–0.842 | <0.001 | |
| >65 years | 0.444 | 0.397–0.497 | <0.001 | 0.507 | 0.450–0.571 | <0.001 | |
| Sex | |||||||
| Female | Reference | ||||||
| Male | 0.972 | 0.926–1.020 | 0.328 | ||||
| Race | |||||||
| American Indian/Alaska Native | Reference | ||||||
| Asian or Pacific Islander | 0.948 | 0.706–1.290 | 0.769 | ||||
| Black | 0.930 | 0.696–1.262 | 0.689 | ||||
| White | 0.711 | 0.535–0.959 | 0.054 | ||||
| Histological grade | |||||||
| Grade I | Reference | Reference | |||||
| Grade II | 1.845 | 1.699–2.006 | <0.001 | 1.627 | 1.495–1.773 | <0.001 | |
| Grade III | 4.011 | 3.621–4.446 | <0.001 | 3.596 | 3.235–4.000 | <0.001 | |
| Grade IV | 3.887 | 3.151–4.776 | <0.001 | 3.519 | 2.830–4.359 | <0.001 | |
| Histological type | |||||||
| Adenocarcinoma | Reference | Reference | |||||
| Mucinous adenocarcinoma | 1.290 | 1.173–1.418 | <0.001 | 1.255 | 1.135–1.385 | <0.001 | |
| Signet-ring cell carcinoma | 2.801 | 1.953–3.972 | <0.001 | 1.481 | 1.016–2.136 | 0.082 | |
| T classification | |||||||
| T1 | Reference | Reference | |||||
| T2 | 1.860 | 1.763–1.963 | <0.001 | 1.763 | 1.666–1.867 | <0.001 | |
| M classification | |||||||
| M0 | Reference | Reference | |||||
| M1 | 8.491 | 7.425–9.724 | <0.001 | 7.318 | 6.373–8.414 | <0.001 | |
| Tumor size | |||||||
| <50 mm | Reference | Reference | |||||
| ≥50 mm | 1.333 | 1.250–1.420 | <0.001 | 1.076 | 1.004–1.152 | 0.080 | |
| Tumor location | |||||||
| Ascending colon | Reference | Reference | |||||
| Cecum | 1.273 | 1.184–1.371 | <0.001 | 1.206 | 1.118–1.301 | <0.001 | |
| Descending colon | 1.300 | 1.152–1.465 | <0.001 | 1.258 | 1.109–1.424 | <0.01 | |
| Hepatic flexure | 1.008 | 0.877–1.155 | 0.927 | 0.938 | 0.812–1.079 | 0.457 | |
| Sigmoid colon | 1.702 | 1.589–1.824 | <0.001 | 1.666 | 1.550–1.792 | <0.001 | |
| Splenic flexure | 1.079 | 0.905–1.279 | 0.469 | 1.034 | 0.863–1.232 | 0.756 | |
| Transverse colon | 0.977 | 0.878–1.086 | 0.718 | 0.975 | 0.874–1.087 | 0.703 | |
LNM, lymph node metastasis; OR, odds ratio; CI, confidence interval.
Development and validation of a prediction model for LNM in early-stage colon cancer
The independent risk factors, such as patient age, histologic grade, pathologic type, T stage, M stage, and tumour location, were used to develop a diagnostic prediction model graphically presented as a nomogram (Figure 1). The corresponding score for each risk factor was initially calculated according to patient-specific information and then the overall risk factors were summed to give a total score. The probabilistic risk of LNM for a patient was predicted by projecting the total score of the patient’s risk factors onto the corresponding risk axis of LNM. The ROC curves of the prediction model for diagnosis were plotted using R, which had an AUC of 0.6714 [95% confidence interval (CI): 0.6621–0.6806] for the training set and 0.6567 (95% CI: 0.6422–0.6712) for the validation set. This suggested that the prediction model had high diagnostic and discriminatory power for either the training set or the validation set, with a C-index of 0.6714 for the training set and 0.6567 for the validation set (Figure 2A,2B).
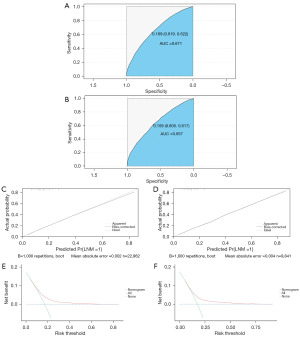
The calibration curves for the training and validation sets confirmed that the prediction model demonstrated good agreement between observed and predicted probabilities (Figure 2C,2D). The DCA showed that the diagnostic prediction model developed in this study had good clinical utility, confirming that the nomogram can be used as an accurate tool to assess the risk of LNM in patients with early-stage colon cancer (Figure 2E,2F).
Discussion
With the development of colonoscopy techniques, low-risk colorectal cancer confined to the mucosa and submucosa can be treated by endoscopic resection. Patients with colorectal cancer at clinical stage T1 can be treated by transanal endoscopic resection, surgery, or a combination of both, all of which generally lead to a good prognosis. Moreover, expanding the current conventional indications for the endoscopic treatment of patients with stage T1 colorectal cancer may not be safe. Adequate surgical resection plus lymph node dissection is recommended for patients with stage T1 colorectal cancer who do not meet the current indications (14). The cause of death in patients with colon cancer is usually distant metastasis of the tumour, but it has been documented that in colon cancer, LNM precedes distant metastasis (2). One study reported that an increase in the number of lymph nodes assessed is associated with an increase in survival rate. Thus, the assessment of lymph nodes is important for both the prognosis and treatment of colon cancer (12). Another study reported on the use of a nomogram to predict the recurrence of colon cancer based on the number of lymph nodes, lymphovascular infiltration, and other risk factors (15). Nomograms have become a common prognostic tool in oncology (16), attributed to their intuitive and easy to use design. Given this context, we aimed to develop a new type of nomogram for predicting LNM in early-stage colon cancer to provide a reference for clinical treatment decision-making. A previous study developed a prediction model to predict LNM in stage T1 colon cancer using five risk factors—tumour location, age, tumour size, degree of tumour differentiation, and carcinoembryonic antigen (CEA) level—and evaluated the prediction model using ROC and calibration curve analysis, but did not use the DCA method to evaluate the clinical utility of the prediction model (17). In another study, researchers used radiomic features, CT-reported lymph node status, and clinical risk factors to develop a radiomic nomogram, which is easy to use for individualized prediction of LNM in patients with preoperative colorectal cancer (18). In the present study, DCA was used to evaluate the clinical utility of the diagnostic prediction model. DCA is a method for evaluating predictive models and diagnostic tests, which Vickers and Elkin introduced in a 2006 publication in Medical Decision Making (19). DCA enables the calculation of the clinical “net benefit” of one or more predictive models or diagnostic tests, which is different from accuracy metrics such as “discrimination” and “calibration” because it incorporates the consequences of the decisions made based on a model or test. The AUC in this paper, although not exceptionally high, aligns consistently with several prior studies. Moreover, the DCA demonstrated good clinical utility in the proper range.
In this retrospective study, patients with stage pT1 or pT2 colon cancer who had undergone surgical treatment were selected as the participants, and a large number of cases and related clinical data were retrieved by screening the U.S. SEER database. A randomization method was applied to divide the data set into a training set and a validation set, with the aim of using a large number of patients’ data to develop and validate a model that can quickly and intuitively assess LNM in early-stage colon cancer. Moreover, the risk factors incorporated into the model were reliable, and their data were readily available. The reason for using these risk factors is that, in practice, easily obtaining risk factor data in a clinical setting makes it easier to predict the risk of LNM in patients. In the present study, age, histologic grade, pathologic type, T stage, M stage, and tumour location were used as risk factors to develop a diagnostic prediction model. Modelling results showed that old age was a significant protective factor for LNM, with the risk of LNM decreasing with increasing age until 65 years. The risk of metastasis reached its lowest level above 65 years of age. This was consistent with a previous finding that younger patients with colorectal cancer tended to have a higher risk of LNM than older patients (20). It seems the old age plays a protective role for LNM in patients with T1 or T2 colon cancer (21). The present study also observed that both the T and M stages of the tumour were risk factors for LNM and that among various pathologic types, signet-ring cell carcinoma had the highest risk of LNM compared to mucinous and non-mucinous adenocarcinoma. However, another recent study found that compared to other pathologic types, the mucinous adenocarcinoma was related to the highest risk of LNM in the predictive model of patients with T1 colon cancer (22). In our study, the risk of LNM was also the highest in patients with sigmoid colon cancer among all tumour locations, all of which were consistent with existing studies. Multivariate analysis showed that tumour size was not an independent risk factor for LNM and, therefore, was not included in the final prediction model. However, a preceding study found that a large tumour implied a higher risk of LNM (17). In addition, it was observed that the risk of LNM in patients with T1 or T2 colon cancer largely tended to increase with increasing histologic grade. It is generally accepted in clinical practice that less differentiated tumours and more malignant tumours increase the risk of LNM. However, the present multivariate analysis and nomogram demonstrated that grade III [odds ratio (OR): 3.596] had a higher risk of LNM than grade IV (OR: 3.519), which aligned with the finding of a recent study on early-onset colorectal cancer (23). It was contrary to the result of a previous study, which showed that a worse pathologic grade indicated a higher risk of LNM (17). This suggested that the degree of differentiation does not always correlate inversely with the risk of LNM. The underlying reason for this exception can be explored using a larger number of cases or by performing fundamental experimental analysis. In conclusion, if a patient has a low-risk probability of LNM based on our model, the patient can be considered for endoscopic dissection. It avoids unnecessary major surgery caused by operative trauma and decreases hospitalization expense.
The present study has certain limitations. Firstly, it did not consider external validation of the prediction model. Secondly, we failed to use genetic markers and pathological data which including mismatch repair deficiency (dMMR), budding and other parameters for model construction, because the SEER database lacks relevant data. Thirdly, the validation set was derived from the same SEER dataset as the training set, likely leading to a spill over effect in the model. To address this problem, external validation using patient data from other large clinical centres must be performed, suggesting a need to use a multi-centre dataset with a large sample size for external validation. Unfortunately, because the discrepancy between clinical and pathological staging, it may cause a problem that the model in our study clinically unusable. To deal with this, the T staging and distant metastasis can be evaluated by routine enhanced magnetic resonance imaging or CT before surgery to maximize the consistency of clinical and pathological staging (24). Furthermore, the SEER database lacks clinical staging information, which may introduce bias if patients with discrepancies between clinical and pathological staging are not excluded (25). In the future, we would like to bring in routine genetic testing for colorectal cancer patients. A diagnostic prediction model developed using clinical risk factors and genetic markers would have a high predictive power for the risk of LNM in patients with colon cancer.
Conclusions
In conclusion, the incidence of LNM in patients with pT1 and pT2 colon cancer was 17.11% according to our study, which has usually been underestimated in the past. The nomogram developed in our study can objectively and accurately predict the individual risk of LNM and help clinicians to optimize clinical decisions, such as endoscopic resection, which can improve the quality of life more so than radical operation for patients with clinically staged T1 or T2 colon cancer.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1451/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1451/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1451/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ulintz PJ, Greenson JK, Wu R, et al. Lymph Node Metastases in Colon Cancer Are Polyclonal. Clin Cancer Res 2018;24:2214-24. [Crossref] [PubMed]
- Edge SB. American joint committee on cancer. In: AJCC Cancer Staging Manual. Cham: Springer; 2010.
- Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145-64. [Crossref] [PubMed]
- Wada H, Shiozawa M, Katayama K, et al. Systematic review and meta-analysis of histopathological predictive factors for lymph node metastasis in T1 colorectal cancer. J Gastroenterol 2015;50:727-34. [Crossref] [PubMed]
- Fields AC, Lu P, Hu F, et al. Lymph Node Positivity in T1/T2 Rectal Cancer: a Word of Caution in an Era of Increased Incidence and Changing Biology for Rectal Cancer. J Gastrointest Surg 2021;25:1029-35. [Crossref] [PubMed]
- Xiong X, Wang C, Wang B, et al. Can transanal endoscopic microsurgery effectively treat T1 or T2 rectal cancer?A systematic review and meta-analysis. Surg Oncol 2021;37:101561. [Crossref] [PubMed]
- Garcia-Aguilar J, Renfro LA, Chow OS, et al. Organ preservation for clinical T2N0 distal rectal cancer using neoadjuvant chemoradiotherapy and local excision (ACOSOG Z6041): results of an open-label, single-arm, multi-institutional, phase 2 trial. Lancet Oncol 2015;16:1537-46. [Crossref] [PubMed]
- Fan Y, Wang Y, He L, et al. Clinical features of patients with HER2-positive breast cancer and development of a nomogram for predicting survival. ESMO Open 2021;6:100232. [Crossref] [PubMed]
- Ou YC, Chang KH, Tung MC, et al. Building a Nomogram for Prediction of Prostate Cancer in Patients With Preoperatively Suspected Prostate Cancer. Anticancer Res 2020;40:2995-3002. [Crossref] [PubMed]
- Jeong SH, Kim RB, Park SY, et al. Nomogram for predicting gastric cancer recurrence using biomarker gene expression. Eur J Surg Oncol 2020;46:195-201. [Crossref] [PubMed]
- Chang GJ, Rodriguez-Bigas MA, Skibber JM, et al. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst 2007;99:433-41. [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
- Kim B, Kim EH, Park SJ, et al. The risk of lymph node metastasis makes it unsafe to expand the conventional indications for endoscopic treatment of T1 colorectal cancer: A retrospective study of 428 patients. Medicine (Baltimore) 2016;95:e4373. [Crossref] [PubMed]
- Weiser MR, Landmann RG, Kattan MW, et al. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol 2008;26:380-5. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Elkin EB, et al. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53. [Crossref] [PubMed]
- Hu DY, Cao B, Li SH, et al. Incidence, risk factors, and a predictive model for lymph node metastasis of submucosal (T1) colon cancer: A population-based study. J Dig Dis 2019;20:288-93. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. [Crossref] [PubMed]
- Ghimire B, Singh YP, Kurlberg G, et al. Comparison of Stage and Lymph Node Ratio in Young and Older Patients with Colorectal Cancer Operated in a Tertiary Hospital in Nepal. J Nepal Health Res Counc 2018;16:89-92. [Crossref] [PubMed]
- Meyer JE, Cohen SJ, Ruth KJ, et al. Young Age Increases Risk of Lymph Node Positivity in Early-Stage Rectal Cancer. J Natl Cancer Inst 2016;108:djv284. [Crossref] [PubMed]
- Xiong X, Wang C, Cao J, et al. Lymph node metastasis in T1-2 colorectal cancer: a population-based study. Int J Colorectal Dis 2023;38:94. [Crossref] [PubMed]
- Liu Y, Sun Z, Guo Y, et al. Construction and validation of a nomogram of risk factors and cancer-specific survival prognosis for combined lymphatic metastases in patients with early-onset colorectal cancer. Int J Colorectal Dis 2023;38:128. [Crossref] [PubMed]
- Kijima S, Sasaki T, Nagata K, et al. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J Gastroenterol 2014;20:16964-75. [Crossref] [PubMed]
- Liu X, Sha L, Huang C, et al. A nomogram prediction model for lymph node metastasis risk after neoadjuvant chemoradiotherapy in rectal cancer patients based on SEER database. Front Oncol 2023;13:1098087. [Crossref] [PubMed]



