
Overexpression of KLHL22 correlates with poor prognosis in patients with triple-negative breast cancer
Highlight box
Key findings
• We report that the Kelch-like family member 22 (KLHL22) protein can serve as a prognostic marker for triple-negative breast cancer (TNBC).
What is known and what is new?
• TNBC has biological characteristics that make it prone to metastasis and recurrence.
• KLHL22 can predict the prognosis of TNBC.
What is the implication, and what should change now?
• Further research is needed on the role of KLHL22 in TNBC.
Introduction
According to the 2020 Global Cancer Statistics Report, an estimated 19.3 million new cancer cases and nearly 10.0 million cancer deaths had occurred worldwide. Female breast cancer has surpassed lung cancer as the most commonly diagnosed cancer and is now ranked as the fifth leading cause of cancer mortality worldwide (1). Triple-negative breast cancer (TNBC) lacks human epidermal growth factor receptor 2 gene amplification and hormone receptor expression and is a subtype of breast tumor that accounts for approximately a quarter of newly diagnosed breast tumors (2). TNBC generally has a poor prognosis, high systemic recurrence rate, and refractoriness to conventional therapy, regardless of the choice of adjuvant treatment (3). Therefore, more effective treatment regimens and prognostic indicators are urgently needed (4).
Kelch-like family member 22 (KLHL22) is a BTB (Broad-complex, Tramtrack, and Bric-à-brac) adaptor protein that usually forms a functional cullin-RING E3 ubiquitin ligase complex with scaffold protein cullin 3 (CUL3) and ring-finger RING box protein 1 (5-7). Studies have shown that inactivation of KLHL22 homologs in nematodes can prolong their lifespan as the protein promotes tumorigenesis and aging by activating mechanistic target of rapamycin in complex 1 (mTORC1) signaling (8). In multiple breast cancer cell lines, the protein level of KLHL22 was elevated, and its deletion prevented mTORC1 activation and tumorigenesis (8).
However, studies on KLHL22 in the context of TNBC are rare, and the clinical prognostic factors of KLHL22 in patients with breast cancer remain unclear. In this study, we explored KLHL22 protein expression in TNBC via immunohistochemistry (IHC) and assessed its relationship with TNBC prognosis using statistical analysis methods. We present this article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-654/rc).
Methods
Patients and tissue specimens
In this retrospective study, 146 patients with TNBC who underwent surgical treatment between 2005 and 2013 at Sun Yat-sen University Cancer Center (Guangzhou, China) were included. The criteria for case selection were based on the pathological diagnosis of TNBC and complete follow-up data were included. Among the patients with TNBC, 75 (51.4%) were aged ≤49 years and 71 (48.6%) were aged >49 years. In these cases, 112 patients (76.7%) were diagnosed at the early stages (I and II), and the remaining 34 (23.3%) were diagnosed at the late stages (III and IV). Table 1 details the clinicopathological features (age, tumor size, Ki67 level, P53 expression, clinical stage, and relapse) of the patients. Tumor staging was defined according to the tumor node metastasis classification system proposed by the American Joint Committee on Cancer/International Union Against Cancer. Tumor differentiation was based on the Edmondson and Steiner criteria.
Table 1
| Variable | All cases | Expression KLHL22, n (%) | P value | |
|---|---|---|---|---|
| Low expression | High expression | |||
| Age (year) | 0.261 | |||
| ≤49 | 75 | 46 (61.3) | 29 (38.7) | |
| >49 | 71 | 37 (52.1) | 34 (47.9) | |
| Tumor size (cm) | 0.826 | |||
| ≤2.5 | 78 | 45 (57.7) | 33 (42.3) | |
| >2.5 | 68 | 38 (55.9) | 30 (44.1) | |
| pT | 0.017 | |||
| T1+2 | 127 | 77 (60.6) | 50 (39.4) | |
| T3+4 | 19 | 6 (31.6) | 13 (68.4) | |
| pN | 0.016 | |||
| N0 | 77 | 51 (66.2) | 26 (33.8) | |
| N1 | 69 | 32 (46.4) | 37 (53.6) | |
| pM | 0.192 | |||
| M0 | 142 | 82 (57.7) | 60 (42.3) | |
| M1 | 4 | 1 (25.0) | 3 (75.0) | |
| Clinical stage | 0.004 | |||
| I+II | 112 | 71 (63.4) | 41 (36.6) | |
| III+IV | 34 | 12 (35.3) | 22 (64.7) | |
| Ki67 | 0.654 | |||
| ≤15 | 61 | 36 (59.0) | 25 (41.0) | |
| >15 | 85 | 47 (55.3) | 38 (44.7) | |
| P53 | 0.485 | |||
| Yes | 95 | 56 (58.9) | 39 (41.1) | |
| No | 51 | 27 (52.9) | 24 (47.1) | |
| Relapse | 0.001 | |||
| Yes | 16 | 3 (18.8) | 13 (81.3) | |
| No | 130 | 80 (61.5) | 50 (38.5) | |
KLHL22, Kelch-like family member 22; TNBC, triple-negative breast cancer.
IHC
Immunohistochemical staining of KLHL22 was performed using the standard EnVision procedure. First, we sequentially cut the paraffin-embedded tissue blocks into 3 µm thick sections and then dried and deparaffinized the slides with the tissue flakes in xylene. Next, the slides were rehydrated through graded alcohol and immersed in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity. After that, the antigen was retrieved by pressure cooking for 3 min in an ethylenediaminetetraacetic acid pH 9.0 antigen retrieval solution. To reduce nonspecific reactions, the slides were incubated with 5% bovine serum albumin for 15 min, after which they were incubated with the mouse anti-KLHL22 monoclonal antibody (1:500 dilution; abs132394; Absin Biotechnology Company, Shanghai, China) for 50 min at 37 ℃. The slides were then incubated with the secondary antibody (Envision, k5007; Dako, Denmark) for 30 min at 37 ℃, followed by staining with 3,3-diaminobenzidine. Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. For the negative control, the primary antibody was replaced with normal mouse immunoglobulin G (IgG).
IHC evaluation
Positive KLHL22 staining was mostly observed in the cytoplasm and were brown or yellow in color. The immunoreactivity score was evaluated by positive cell number and positive intensity score. We first evaluated the percentage of positive tumor cells by taking five fields per slide to count the percentage in 5% increments from 0 to 100%, with 0 indicating negative staining. A positive intensity score was then defined in four degrees as follows: negative [0], weak [1], moderate [2], and strong [3]. Total scores were obtained from the intensity and proportion (0–300 scores). Then, we used the receiver operating characteristic (ROC) curve to determine the cut-off value for KLHL22 expression level in TNBC. Three independent senior pathologists who were blinded to the clinicopathological data of the patients scored KLHL22 expression. All analyses were performed by at least two pathologists in a single laboratory. The value was selected when at least two were in agreement with the results of the diagnosis. If they had different opinions, the diagnosis would be repeated and discussed to reach a consensus. The diagnostic coincidence rate was approximately 75%, which proves that this method has high repeatability.
Statistical analysis
All statistical analyses were performed using SPSS software (version 21.0; SPSS Inc., Chicago, IL, USA). ROC analysis was used to determine the cut-off value for KLHL22. The Chi-squared test was used to evaluate the relationship between the clinicopathological characteristics of patients with TNBC and KLHL22 protein levels. We used the Kaplan-Meier method with the log-rank test to evaluate the survival of patients with TNBC for univariate analysis. The independent prognostic factors were identified using Cox regression analysis. Statistical significance for two-tailed hypothesis was set at P value <0.05.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center (No. SZR2021-053). Written informed consent for participation was not required due to the retrospective nature of this study. All samples were anonymized.
Results
Cut-off value for KLHL22 expression
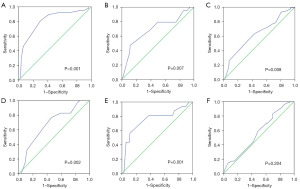
Immunohistochemical analysis showed that in TNBC, KLHL22 was mainly expressed in the cytoplasm (Figure 1). The cut-off score for positive KLHL22 expression was determined using ROC curves. According to previously reported methods, the point with maximum specificity and sensitivity was selected as the cut-off point (9). The sensitivity and specificity for each outcome were plotted as follows: survival outcome (Figure 2A), primary tumor (pT) status (Figure 2B), regional lymph node (pN) status (Figure 2C), clinical stage (Figure 2D), relapse (Figure 2E), and tumor size (Figure 2F). Based on the ROC curve analysis, the cut-off value for the expression of KLHL22 protein was 190 (P<0.001). Using this cut-off, a high level of KLHL22 was detected in 56.85% (83/146) of the TNBC samples.
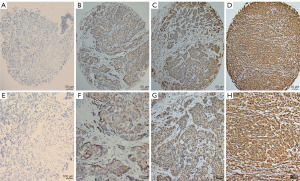
Association between KLHL22 expression and clinicopathologic features of patients with TNBC
Chi-squared analysis demonstrated that KLHL22 expression was significantly correlated with pT and pN status, clinical stage, and relapse (Table 1, P<0.05). There was no significant correlation between KLHL22 expression and other clinicopathological features, such as patient age, tumor size, metastasis (pM) status, and P53 and Ki67 expression (Table 1, P>0.05).
Correlation between KLHL22 expression status, clinicopathological characteristics, and patients survival
Univariate survival analysis revealed a significant effect of clinicopathological prognostic parameters, such as pT (P=0.001), pN (P<0.001), clinical stage (P<0.001), and relapse (P<0.001), on prognosis. The results also demonstrated that KLHL22 expression (P<0.001) could also be used as an important factor in evaluating the prognosis of TNBC (Table 2). Kaplan-Meier survival analysis (Figure 3) showed that the mean survival time of the low KLHL22 group was significantly longer than that of the high KLHL22 group (147.93 vs. 90.1 months, P<0.001; Figure 3A).
Table 2
| Variable | All cases | Mean survival (months) | χ2 | P value |
|---|---|---|---|---|
| Age (year) | 0.1 | 0.752 | ||
| ≤49 | 75 | 126.01 | ||
| >49 | 71 | 123.31 | ||
| Tumor size (cm) | 1.22 | 0.269 | ||
| ≤2.5 | 78 | 129.64 | ||
| >2.5 | 68 | 119.03 | ||
| pT | 10.103 | 0.001 | ||
| T1+2 | 127 | 129.75 | ||
| T3+4 | 19 | 85.36 | ||
| pN | 21.471 | <0.001 | ||
| N0 | 77 | 145.06 | ||
| N1 | 69 | 102.95 | ||
| pM | 1.909 | 0.167 | ||
| M0 | 142 | 125.67 | ||
| M1 | 4 | 68.5 | ||
| Clinical stage | 34.255 | <0.001 | ||
| I+II | 112 | 138.43 | ||
| III+IV | 34 | 77.37 | ||
| Ki67 | 0.464 | 0.496 | ||
| ≤15 | 61 | 128.52 | ||
| >15 | 85 | 108.96 | ||
| P53 | 2.677 | 0.102 | ||
| Yes | 95 | 11.75 | ||
| No | 51 | 135.82 | ||
| Relapse | 45.469 | <0.001 | ||
| Yes | 16 | 49.19 | ||
| No | 130 | 133.91 | ||
| KLHL22 | 41.476 | <0.001 | ||
| High | 63 | 90.1 | ||
| Low | 83 | 147.93 |
TNBC, triple-negative breast cancer.
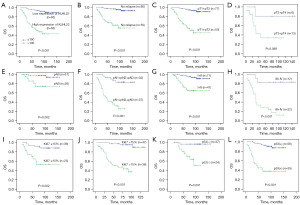
We further evaluated the relationship between KLHL22 protein expression and prognosis of patients with TNBC in different clinicopathological subgroups. The results showed that high KLHL22 expression was associated with poor prognosis in the no relapse (P<0.001; Figure 3B), pT1+pT2 (P<0.001; Figure 3C), pN0 (P=0.002; Figure 3E), pN1+pN2+pN3 (P<0.001; Figure 3F), stage I+II (P<0.001; Figure 3G), stage III+IV (P<0.001; Figure 3H), Ki67 ≤15% (P=0.002; Figure 3I), Ki67 >15% (P<0.001; Figure 3J), p53(−) (P=0.001; Figure 3K), and p53(+) (P<0.001; Figure 3L) groups. Regardless of the Ki67 expression rate (i.e., >15% or ≤15%), high KLHL22 expression was associated with shorter overall survival. Similarly, regardless of whether p53 was negatively or positively expressed, the group with lower KLHL22 protein expression levels had longer overall survival. However, there was no significant relationship between KLHL22 expression and prognosis in the pT3+pT4 group (P=0.089; Figure 3D). In general, patients with low KLHL22 levels had a longer mean survival than those with high KLHL22 levels. It was associated with no relapse, pT1+pT2, pN0, pN1+pN2+pN3, stage I+II, stage III+IV, and Ki67 and p53 expression (Figure 3).
Independent prognostic factors for patents with TNBC
Multivariate analysis showed that P53 expression, clinical stage, and KLHL22 level were independent prognostic factors for overall survival (P<0.05), whereas clinical stage and KLHL22 level were independent prognostic factors for progression-free survival (P<0.05; Table 3). We performed Cox regression analysis for several factors that showed significant differences in the univariate analysis. As shown in Table 3, high KLHL22 level was an independent prognostic factor associated with poor overall survival [hazard ratio (HR): 10.41; 95% confidence interval (CI): 4.313–25.126; P<0.001] and progression-free survival (HR: 8.493; 95% CI: 2.210–32.642; P=0.002). At the same time, it was also found that P53 expression (P=0.024) and clinical stage (P<0.001) were independent prognostic factors for overall patient survival, while clinical stage (P<0.001) was an independent prognostic factors for progression-free survival (Table 3).
Table 3
| Variable | OS | PFS | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (>49 vs. ≤49) (year) | 0.602 | 0.300–1.207 | 0.152 | 0.52 | 0.185–1.466 | 0.216 | |
| P53 (yes vs. no) | 2.441 | 1.125–5.299 | 0.024 | 0.633 | 0.234–1.714 | 0.368 | |
| Tumor size (>2.5 vs. ≤2.5) (cm) | 0.847 | 0.424–1.691 | 0.637 | 0.74 | 0.254–2.154 | 0.581 | |
| Clinical stage (III+IV vs. I+II) | 4.596 | 2.260–9.345 | <0.001 | 3.093 | 1.062–9.008 | 0.038 | |
| KLHL22 (high vs. low) | 10.41 | 4.313–25.126 | <0.001 | 8.493 | 2.210–32.642 | 0.002 | |
TNBC, triple-negative breast cancer; OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval.
Discussion
As a member of the KLHL family, KLHL22 encodes a group of proteins that typically contain a BTB/POZ domain and is most visible as a diffuse cytoplasmic signal (7,10). As an adaptor of the CUL3-based E3 ligase, KLHL22 activates amino acid-dependent mTORC1 signaling to promote aging and tumorigenesis (8). KLHL22 can also affect translational modification through the role of CUL3-based ubiquitin ligase, thereby affecting cell function (6,11,12). KLHL22 mRNA levels are elevated in prostate cancer, breast cancer, and melanoma, according to the Oncomine database. KLHL22 protein levels are elevated in multiple breast cancer cell lines and breast tumor tissues of patients, and KLHL22 deletion prevents mTORC1 activation and tumorigenesis in breast cancer cells (8). However, the upstream molecular mechanism of KLHL22 is not thoroughly studied, and its role in tumor development remains unclear.
In this study, a high level of KLHL22 was detected in the TNBC samples. We also found a significant relationship between KLHL22 expression and clinicopathological factors, such as pT and pN status, clinical stage, and relapse. Among patients with TNBC, those with low KLHL22 levels had longer mean survival than those with high KLHL22 levels.
Ki67 expression is related to the proliferation and growth of many tumor cells and is used as a proliferation marker in routine pathological examinations (13). Although the value of Ki67 in breast cancer has been controversial, recent guidelines have included it in pathologic tests as a biological marker (14). Ki67 has also been proposed as a biological tumor and prognostic marker in breast cancer (15). In this study, we detected KLHL22 and Ki67 expression in TNBC using IHC and found that high KLHL22 expression was significantly associated with poor prognosis in the Ki67 ≤15% and Ki67 >15% groups. This provides evidence for its potential role in TNBC.
In addition, we found that p53 expression was an independent prognostic factor for the overall survival of patients with TNBC. Moreover, a high level of KLHL22 was a prognostic factor in patients with p53(+) and p53(−) expression. However, the relationship between KLHL22 and p53 remains unclear. Jeong found that Daxx-mediated repression of ETS1- and p53-dependent transcription was reversed after expression of SPOP with CUL3 (11), which might explain the relationship between p53 expression and KLHL22. Li et al. (16) showed that p53 is a candidate gene for poor prognosis in patients with TNBC. This is consistent with the results of our study, which showed that p53 expression was an independent prognostic factor for the overall survival of patients with TNBC.
Chen et al. found that the CUL3-KLHL22 E3 ubiquitin ligase promotes the degradation of DEP domain containing 5 (DEPDC5), an essential subunit of GTPase-activating protein (GAP) Activity TOward Rags 1 (GATOR1), and K48-linked polyubiquitination in response to amino acids. The GATOR1 complex, which is composed of DEPDC5, NPRL3, and NPRL2, exhibits GAP activity to inactivate RAG GTPases under amino acid-deficient conditions (8). RAG GTPases can recruit mTORC1 to lysosomes in response to amino acids (17). mTORC1 is a primary regulator of cell growth (18) that is unregulated and is associated with cancer. Deregulation of mTORC1 has been associated with carcinogenesis (19,20), and this explains how KLHL22 activates amino acid-dependent mTORC1 signaling to promote tumors. This is consistent with our finding that KLHL22 is an oncoprotein in TNBC. In addition, it has been reported that KLHL22 promoted breast tumorigenesis by activating mTORC1 signaling (8). Liu et al. (21) found that KLHL22 expression was upregulated in human malignant melanoma tissues and demonstrated that its overexpression promoted malignant melanoma cell growth in vitro. These studies concluded that KLHL22 functions as an oncogene, and these results are consistent with ours. Overexpression of KLHL22 by tumor cells resulted in a poor prognosis for these patients. The overall and progression-free survival rates were significantly longer in the low KLHL22 group than in the high KLHL22 group. Overall, KLHL22 could be used as an independent prognostic factor to predict the overall and progression-free survival of patients with TNBC, as evidenced by our clinical data and expression studies. However, a study by Song et al. (22) found that KLHL22 protein levels in clinicopathological tissue samples of patients were lower than those in normal tissues, and they further explained that KLHL22 acts as a tumor suppressor and regulates proliferation and epithelial-mesenchymal transition through the Wnt/β-catenin signaling pathway in colorectal cancer cells. In addition, Zhou et al. (23) revealed that KLHL22 plays a crucial role in maintaining the programmed cell death protein 1 (PD-1) expression balance and preventing excessive suppression of T cells. KLHL22 deficiency inhibits the antitumor response of T cells and promotes tumor progression through an excessive accumulation of PD-1. KLHL22 may function as a tumor suppressor gene; however, its role in tumor immunity and tumorigenesis warrants further study. Furthermore, this study is limited to single-center data and needs to be expanded to a multi-center validation analysis. Additionally, the molecular mechanism of KLHL22 in breast cancer needs to be further explored.
Conclusions
In summary, detection of KLHL22 expression via IHC is an effective tool to identify the prognosis of patients with TNBC. Our study also provides evidence that KLHL22 may be a potential therapeutic target for TNBC.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-654/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-654/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-654/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-654/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center (No. SZR2021-053). Written informed consent for participation was not required due to the retrospective nature of this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Borri F, Granaglia A. Pathology of triple negative breast cancer. Semin Cancer Biol 2021;72:136-45. [Crossref] [PubMed]
- Jovanović B, Mayer IA, Mayer EL, et al. A Randomized Phase II Neoadjuvant Study of Cisplatin, Paclitaxel With or Without Everolimus in Patients with Stage II/III Triple-Negative Breast Cancer (TNBC): Responses and Long-term Outcome Correlated with Increased Frequency of DNA Damage Response Gene Mutations, TNBC Subtype, AR Status, and Ki67. Clin Cancer Res 2017;23:4035-45. [Crossref] [PubMed]
- Jia H, Truica CI, Wang B, et al. Immunotherapy for triple-negative breast cancer: Existing challenges and exciting prospects. Drug Resist Updat 2017;32:1-15. [Crossref] [PubMed]
- Dhanoa BS, Cogliati T, Satish AG, et al. Update on the Kelch-like (KLHL) gene family. Hum Genomics 2013;7:13. [Crossref] [PubMed]
- Maerki S, Olma MH, Staubli T, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol 2009;187:791-800. [Crossref] [PubMed]
- Beck J, Maerki S, Posch M, et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat Cell Biol 2013;15:430-9. [Crossref] [PubMed]
- Chen J, Ou Y, Yang Y, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 2018;557:585-9. [Crossref] [PubMed]
- Cai MY, Zhang B, He WP, et al. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci 2010;101:1543-9. [Crossref] [PubMed]
- Beck J, Peter M. Regulating PLK1 dynamics by Cullin3/KLHL22-mediated ubiquitylation. Cell Cycle 2013;12:2528-9. [Crossref] [PubMed]
- Kwon JE, La M, Oh KH, et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem 2006;281:12664-72. [Crossref] [PubMed]
- Chen RH. Cullin 3 and Its Role in Tumorigenesis. Adv Exp Med Biol 2020;1217:187-210. [Crossref] [PubMed]
- Li LT, Jiang G, Chen Q, et al. Ki67 is a promising molecular target in the diagnosis of cancer Mol Med Rep 2015;11:1566-72. (review). [Crossref] [PubMed]
- Yerushalmi R, Woods R, Ravdin PM, et al. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 2010;11:174-83. [Crossref] [PubMed]
- Zhu H, Doğan BE. American Joint Committee on Cancer's Staging System for Breast Cancer, Eighth Edition: Summary for Clinicians. Eur J Breast Health 2021;17:234-8.
- Li JP, Zhang XM, Zhang Z, et al. Association of p53 expression with poor prognosis in patients with triple-negative breast invasive ductal carcinoma. Medicine (Baltimore) 2019;98:e15449. [Crossref] [PubMed]
- Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496-501. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011;12:21-35. [Crossref] [PubMed]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 2006;6:729-34. [Crossref] [PubMed]
- Liu XR, Wang W, Li HM. KLHL22 promotes malignant melanoma growth in vitro and in vivo by activating the PI3K/Akt/mTOR signaling pathway. Neoplasma 2020;67:1106-13. [Crossref] [PubMed]
- Song Y, Yuan H, Wang J, et al. KLHL22 Regulates the EMT and Proliferation in Colorectal Cancer Cells in Part via the Wnt/β-Catenin Signaling Pathway. Cancer Manag Res 2020;12:3981-93. [Crossref] [PubMed]
- Zhou XA, Zhou J, Zhao L, et al. KLHL22 maintains PD-1 homeostasis and prevents excessive T cell suppression. Proc Natl Acad Sci U S A 2020;117:28239-50. [Crossref] [PubMed]


